Welcome to CPSS
User can prepare their input file from HERE.
Users can start analysis from HERE ( Single / Paired ).
"Single" means the dataset from one sample, and "Paired" means the datasets from two samples.
News
A updated version of CPSS, CPSS2.0, was online at here. Please feel free to use it. (2017-10-05)
Introduction
Key words: Small RNA deep-sequencing, mircoRNA, piRNA, CPSS
Non-coding small RNAs are RNA molecules that are not translated into proteins, including ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), microRNAs (miRNA), piwi-interacting RNAs (piRNAs) and other RNA species (1). These RNAs are also known as the regulators that carry out a variety of biological functions, have been discovered only 20 years ago (2).
MicroRNAs, first discovered in C. Elegans (3), are short RNA molecules, approximately 22 nucleotides in length. Mature miRNAs are post-transcriptional regulators that bind to complementary sequences on target mRNAs, usually resulting in translational repression, degradation, or both of the target mRNAs. The importance of miRNAs is evidenced by their evolutionary conservation across species and by many cellular pathways in which they are involved, including growth, proliferation, apoptosis, and developmental timing (4).
Piwi-interacting RNAs (piRNAs) are typically ~30 bases long, as a distinct population of piwi-associated small RNAs first discovered in Drosophila. The function of piRNAs is currently unknown, but the homology of PIWI proteins to Argonaute proteins and similarities of piRNAs to microRNAs and short-interfering RNAs suggest that piRNAs play a role in regulatory processes during meiosis, particularly those in spermatogenesis (5).
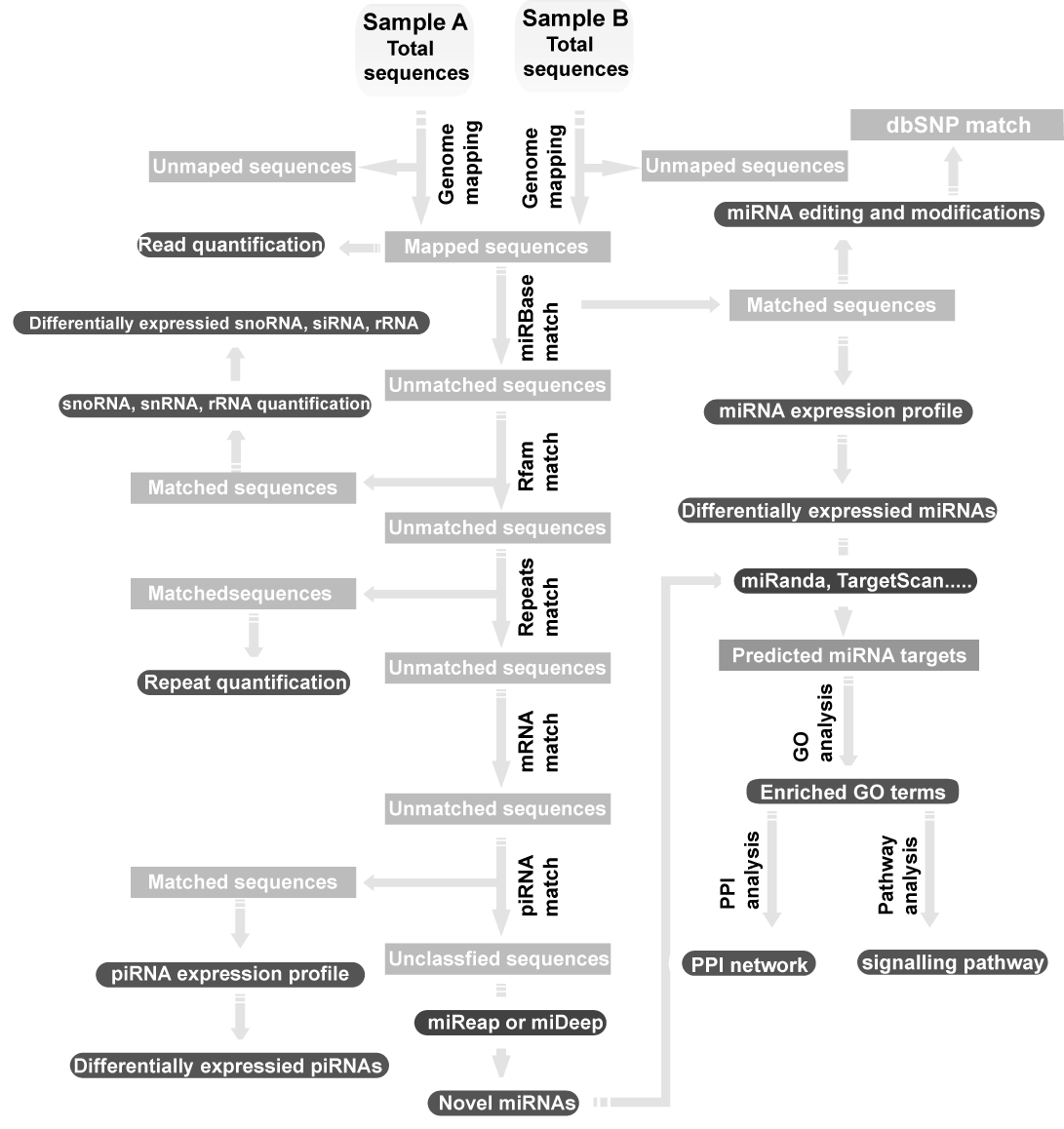
Deep sequencing technologies have found broad applicability in functional genomics research (6). The main advantage of deep sequencing technologies is eliminating the need for in vivo cloning by clonal amplification of spatially separated single molecules using either emulsion PCR (Roche 454 and Applied Biosystems SOLiD) or bridge amplification on a solid surface (Illumina Solexa Genome Analyzer). Deep sequencing has been used extensively for expression profiling and discovery of microRNAs (miRNAs) and other small non-coding RNAs (ncRNAs) in many organisms.
To handle the millions of short sequences from deep sequencing technologies for small RNAs, the researchers are badly in need of a systematic application platform for data analysis. Therefore, a comprehensive and free web server CPSS was developed for small RNA quantification, prediction of novel microRNAs, prediction of microRNA targets, and microRNA functional annotation and prediction.
1. Moazed D. (2009) Small RNAs in transcriptional gene silencing and genome defence. Nature, 457, 413-20.
2. Amaral PP, Mattick JS. (2008) Noncoding RNA in development. Mamm Genome, 19, 454-492.
3. Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 75, 843-854.
4. Carthew, R.W. and Sontheimer, E.J. (2009) Origins and Mechanisms of miRNAs and siRNAs, Cell, 136, 642-655.
5. Aravin AA, Hannon GJ, Brennecke J (2007) The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science, 318(5851), 761-4.
6. Zhou, L., et al. (2011) Small RNA transcriptome investigation based on next-generation sequencing technology. J Genet Genomics. Nov 20, 505-13.