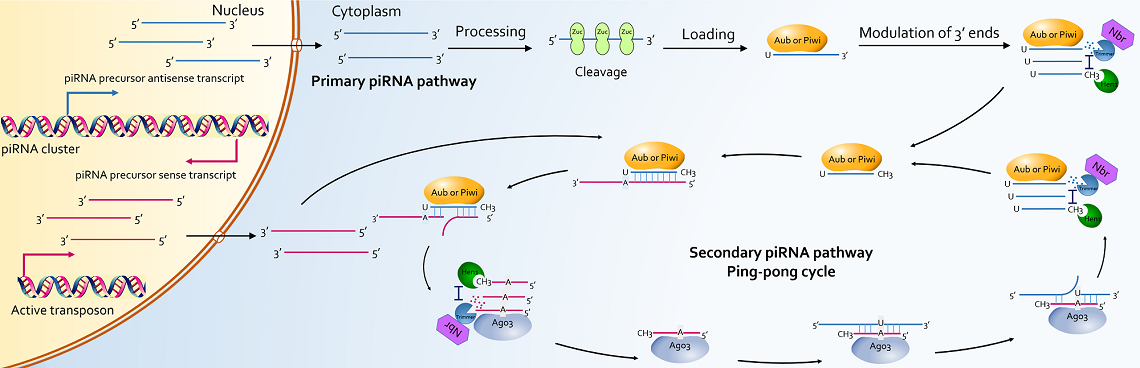
PIWI-interacting RNAs (piRNAs) are essential for transcriptional and post-transcriptional regulation of transposons and coding genes in germlines. With the development of sequencing technologies, length variations of piRNAs have been identified in several species. In Drosophila, AT-chX-1, one prominent testicular piRNA, has multiple isoforms with heterogeneous 3’ ends. Loss function of HENMT1, an enzyme involved in piRNA 3’end modulation, leads to piRNA instability, derepression of TEs and ultimately male sterility in mouse. However, the extent to which, piRNA isoforms are existed, and whether these isoforms are functionally distinct from canonical piRNAs remain uncharacterized. Through data mining from 2,154 datasets of small RNA sequencing data from four species (Homo sapiens, Mus musculus, Danio rerio and Drosophila melanogaster), we have identified 8,749,139 piRNA isoforms from 175,454 canonical piRNAs, and classified them on the basis of variations on 5’ or 3’ end via the alignment of isoforms with canonical sequence. We thus established a database named IsopiRBank. Each isoforms has detailed annotation as follows: normalized expression data, classification, spatiotemporal expression data and genome origin. Users can also select interested isoforms for further analysis, including target prediction, and Enrichment analysis. Taken together, IsopiRBank is an interactive database that aims to present the first integrated resource of piRNA isoforms, and broaden the research of piRNA biology. IsopiRBank can be accessed at http://mcg.ustc.edu.cn/bsc/isopir/index.html without any registration or log in requirement.
1. Siomi, M.C., Sato, K., Pezic, D. and Aravin, A.A. (2011) PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Bio, 12, 246-258.
2. Lau, N.C., Seto, A.G., Kim, J., Kuramochi-Miyagawa, S., Nakano, T., Bartel, D.P. and Kingston, R.E. (2006) Characterization of the piRNA complex from rat testes. Science, 313, 363-367.
3. Carmell, M.A., Girard, A., van de Kant, H.J.G., Bourc'his, D., Bestor, T.H., de Rooij, D.G. and Hannon, G.J. (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell, 12, 503-514.
4. Zheng, K. and Wang, P.J. (2012) Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity. Plos Genet, 8.
5. Klattenhoff, C. and Theurkauf, W. (2008) Biogenesis and germline functions of piRNAs. Development, 135, 3-9.
6. Le Thomas, A., Toth, K.F. and Aravin, A.A. (2014) To be or not to be a piRNA: genomic origin and processing of piRNAs. Genome Biol, 15.
7. Houwing, S., Kamminga, L.M., Berezikov, E., Cronembold, D., Girard, A., van den Elst, H., Filippov, D.V., Blaser, H., Raz, E., Moens, C.B. et al. (2007) A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell, 129, 69-82.
8. Lim, A.K., Tao, L.H. and Kai, T. (2009) Post-transcriptional retroelement surveillance is mediated by piRNAs in Drosophila germline. Mech Develop, 126, S51-S51.
9. Sienski, G., Donertas, D. and Brennecke, J. (2012) Transcriptional Silencing of Transposons by Piwi and Maelstrom and Its Impact on Chromatin State and Gene Expression. Cell, 151, 964-980.
10. Le Thomas, A., Rogers, A.K., Webster, A., Marinov, G.K., Liao, S.E., Perkins, E.M., Hur, J.K., Aravin, A.A. and Toth, K.F. (2013) Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Gene Dev, 27, 390-399.
11. Rouget, C., Papin, C., Boureux, A., Meunier, A.C., Franco, B., Robine, N., Lai, E.C., Pelisson, A. and Simonelig, M. (2010) Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature, 467, 1128-U1144.
12. Gou, L.T., Dai, P., Yang, J.H., Xue, Y.C., Hu, Y.P., Zhou, Y., Kang, J.Y., Wang, X., Li, H.R., Hua, M.M. et al. (2014) Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res, 24, 680-700.
13. Zhang, P., Kang, J.Y., Gou, L.T., Wang, J.J., Xue, Y.C., Skogerboe, G., Dai, P., Huang, D.W., Chen, R.S., Fu, X.D. et al. (2015) MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res, 25, 193-207.
14. Barberan-Soler, S., Fontrodona, L., Ribo, A., Lamm, A.T., Iannone, C., Ceron, J., Lehner, B. and Valcarcel, J. (2014) Co-option of the piRNA Pathway for Germline-Specific Alternative Splicing of C-elegans TOR. Cell Rep, 8, 1609-1616.
15. Zhang, P., Si, X.H., Skogerbo, G., Wang, J.J., Cui, D.Y., Li, Y.X., Sun, X.B., Liu, L., Sun, B.F., Chen, R.S. et al. (2014) piRBase: a web resource assisting piRNA functional study. Database-Oxford.
16. Aravin, A., Gaidatzis, D., Pfeffer, S., Lagos-Quintana, M., Landgraf, P., Iovino, N., Morris, P., Brownstein, M.J., Kuramochi-Miyagawa, S., Nakano, T. et al. (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature, 442, 203-207.
17. Chirn, G.W., Rahman, R., Sytnikova, Y.A., Matts, J.A., Zeng, M., Gerlach, D., Yu, M., Berger, B., Naramura, M., Kile, B.T. et al. (2015) Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals. Plos Genet, 11.
18. Han, B.W., Wang, W., Li, C.J., Weng, Z.P. and Zamore, P.D. (2015) piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science, 348, 817-821.
19. Lim, S.L., Qu, Z.P., Kortschak, R.D., Lawrence, D.M., Geoghegan, J., Hempfling, A.L., Bergmann, M., Goodnow, C.C., Ormandy, C.J., Wong, L. et al. (2015) HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse. Plos Genet, 11.
20. Feltzin, V.L., Khaladkar, M., Abe, M., Parisi, M., Hendriks, G.J., Kim, J. and Bonini, N.M. (2015) The exonuclease Nibbler regulates age-associated traits and modulates piRNA length in Drosophila. Aging Cell, 14, 443-452.
21. Wang, H., Ma, Z., Niu, K., Xiao, Y., Wu, X., Pan, C., Zhao, Y., Wang, K., Zhang, Y. and Liu, N. (2016) Antagonistic roles of Nibbler and Hen1 in modulating piRNA 3' ends in Drosophila. Development, 143, 530-539.
22. Honda, S., Kirino, Y., Maragkakis, M., Alexiou, P., Ohtaki, A., Murali, R., Mourelatos, Z. and Kirino, Y. (2013) Mitochondrial protein BmPAPI modulates the length of mature piRNAs. Rna, 19, 1405-1418.
23. Han, B.W., Hung, J.H., Weng, Z.P., Zamore, P.D. and Ameres, S.L. (2011) The 3 '-to-5 ' Exoribonuclease Nibbler Shapes the 3 ' Ends of MicroRNAs Bound to Drosophila Argonaute1. Curr Biol, 21, 1878-1887.
24. Lakshmi, S.S. and Agrawal, S. (2008) piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res, 36, D173-D177.
25. Rosenkranz, D. and Zischler, H. (2012) proTRAC - a software for probabilistic piRNA cluster detection, visualization and analysis. Bmc Bioinformatics, 13.
26. Jung, I., Park, J.C. and Kim, S. (2014) piClust: A density based piRNA clustering algorithm. Comput Biol Chem, 50, 60-67.
27. Sarkar, A., Maji, R.K., Saha, S. and Ghosh, Z. (2014) piRNAQuest: searching the piRNAome for silencers. Bmc Genomics, 15.
28. Rosenkranz, D. (2016) piRNA cluster database: a web resource for piRNA producing loci. Nucleic Acids Res, 44, D223-D230.
29. Zhang, Y.W., Xu, B., Yang, Y.F., Ban, R.J., Zhang, H., Jiang, X.H., Cooke, H.J., Xue, Y. and Shi, Q.H. (2012) CPSS: a computational platform for the analysis of small RNA deep sequencing data. Bioinformatics, 28, 1925-1927.
30. Yang, Q.L., Hua, J., Wang, L., Xu, B., Zhang, H., Ye, N., Zhang, Z.Q., Yu, D.X., Cooke, H.J., Zhang, Y.W. et al. (2013) MicroRNA and piRNA Profiles in Normal Human Testis Detected by Next Generation Sequencing. Plos One, 8.
31. Neilsen, C.T., Goodall, G.J. and Bracken, C.P. (2012) IsomiRs - the overlooked repertoire in the dynamic microRNAome. Trends Genet, 28, 544-549.
32. Hackenberg, M., Sturm, M., Langenberger, D., Falcon-Perez, J.M. and Aransay, A.M. (2009) miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res, 37, W68-W76.
33. Mani, S.R. and Juliano, C.E. (2013) Untangling the Web: The Diverse Functions of the PIWI/piRNA Pathway. Mol Reprod Dev, 80, 632-664.