Meiosis Online
Key words: Meiosis , Reproductive, Genes/Proteins
Meiosis Online scheme

Abbreviation cKO: conditional knockout; IF: Immunofluorescence; IHC: Immunohistochemistry; TUNNEL: TUNNEL assay; WB: Western blotting; Imaging: Live Cell imaging; KO: Knockout; Microinjection: Microinject antibody or inhibitor; Mutation: Point mutation or other change of the nucleotide sequence of the genome; Overexpression: A gene expression in increased quantity; PCR: Polymerase chain reaction; RNA interference: Down-regulation of gene in vitro; Transgene: Genetic material transferred by genetic engineering techniques; Treatment: Antibody, inhibitor or chemical treatment; ISH: In Situ Hybridization *:experiment varified records.
The Meiosis Online provides the browse, search option and advanced options.
1. Browse You can browse the Meiosis Online database by classification and species group tree . 
2. Search You can input keywords to search the Meiosis Online database. The search fields include MG ID, UniProt Accession, Protein Names, Gene Names, Function in Stage, Function in Sex, Effect on Fecundity, and Methods.
Example: Please click on the Species field in the combo box to search "Homo sapiens". By clicking on the "Submit" button, the related MG proteins will be shown.
3. Advanced Options Four advanced options are provided, including advanced search, BLAST search, orthologous search.
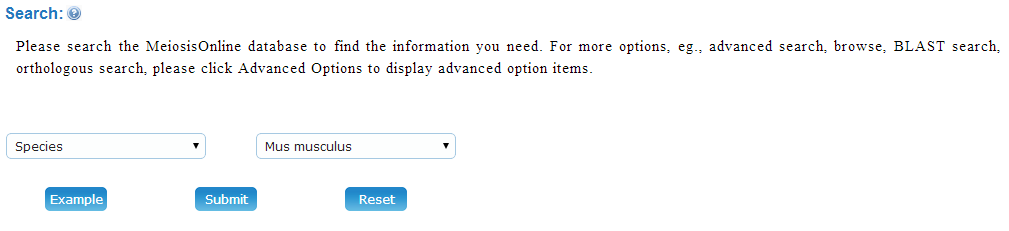
(1) Advanced search: Advanced search allows you to input up to three terms to find the information more specifically. The querying fields can be empty if fewer terms were needed. The three terms could be connected by the following operators:
and: the term following this operator has to be included in the specified field(s).
or: either the preceding or the following term to this operator should occur in the specified field(s).
exclude: If selected, the term following this operator must be not contained in the specified field(s).
Example: You can click on the "Example" button to load an instance, which could search proteins that function in "Male" and "Prophase I" causing infertility. Finalinly, 147 proteins of will find in Meiosis Online.
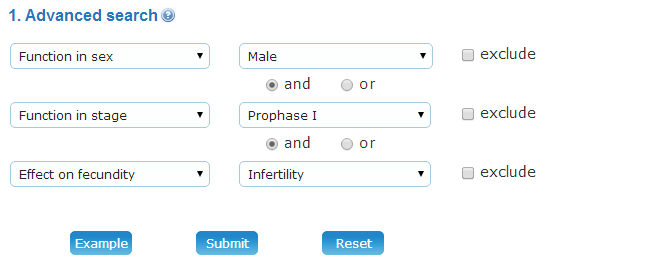
(2) BLAST search: BLAST search could be used to find the specific protein and/or related homologues by sequence alignment. This search-option will help you to find the querying protein accurately and quickly. Only one protein sequence in FASTA format is allowed per time. The E-value threshold could be user-defined. The BLAST results will be parsed and shown by clicking on the "Submit" button.
Example: You can click on the "Example" button to load the protein sequence of mouse Peroxisome proliferator-activated receptor gamma coactivator-related protein 1. By clicking on the "Submit" button, you can find the related homologues in Meiosis Online database.
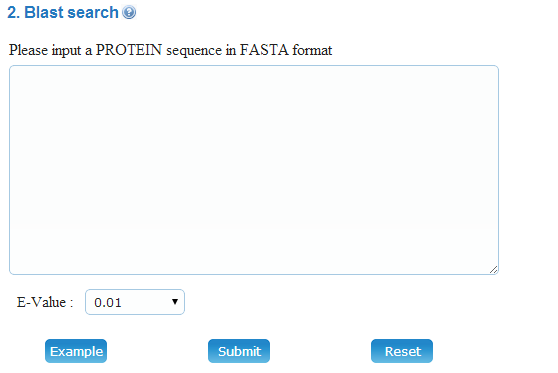
(3) Orthology search: In Meiosis Online database, the pairwise orthologous information was computed with the Panther and InParanoid program (Mi H.et al., 2010; Remm M et al., 2001; Kevin P et al., 2005). Two operators are provided:
(a) Simple: You can input a specified uniport Accession that include in Meiosis Online database. The Identity, E-value and BLAST Score thresholds could be user-defined.
Example: You can click on the "Example" button to browse the ENSMUSG00000022429 (Meiotic recombination protein DMC1/LIM15 homolog) orthologous in Meiosis Online database.
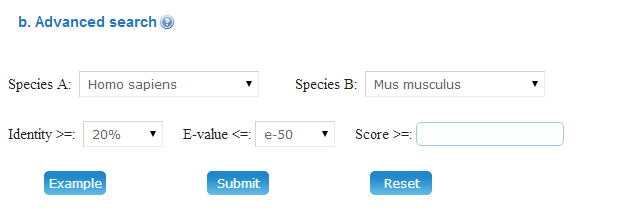
(b) Advanced: Advanced: The pairwise orthologous information could also be viewed in details. The Score (bits), E-value, Identities and Positives from BLAST results are provided.
Example: You can click on the "Example" button to browse the meiosis proteins conserved between Homo spaines and Mus musculus.
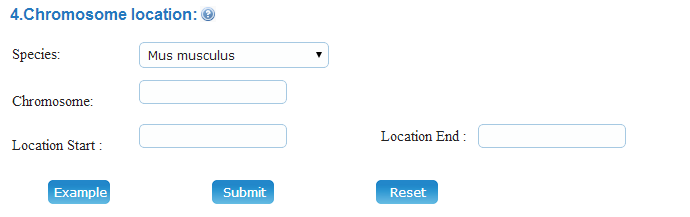
(4) Chromosome Location:In Meiosis Online database, users can search the meiosis-related genes by entry the chromosome location of different species.
Example: You can click on the "Example" button to browse the meiosis genes that locate at 30000000-99999999 in mose chromosome 9.
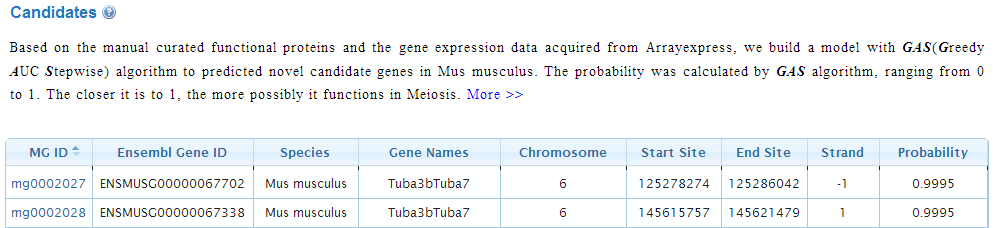
3. Candidates
In Meiosis Online, spermatogenesis-related genome-wide transcriptional data are also collected for data mining. Based on the identified genes and transcriptional data, functional candidate genes which may participate in regulation of spermatogenesis are predicted by the GAS (Greed AUC Stepwise) model. The probability was calculated by GAS algorithm, ranging from 0 to 1. The closer it is to 1, the more possibly it functions in spermatogenesis. Without enough prior information to optimize the critical point. At that point, both the FPR and FNR are relative low according to our simulation study. The users could sort the lsit of predicted genes by click on the "Arrow" button on the side of SG ID, Chromosome, and Probabitity. Detatils of GAS algorithm.
4. Feedback Feedback Users are welcome to review records in Meiosis Online. You can revise the existed records or submit new gene/protein or gene/protein with new function in meiosis. We are grateful to you for your advice!