| Tag | Content | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00000038 |
||||||||||||||||||||||||||||||
UniProt Accession |
|||||||||||||||||||||||||||||||
Theoretical PI |
4.13
|
||||||||||||||||||||||||||||||
Molecular Weight |
27997 Da
|
||||||||||||||||||||||||||||||
Genbank Nucleotide ID |
|||||||||||||||||||||||||||||||
Genbank Protein ID |
|||||||||||||||||||||||||||||||
Gene Name |
asfl-1 |
||||||||||||||||||||||||||||||
Gene Synonyms/Alias |
ORFNames=C03D6.5 |
||||||||||||||||||||||||||||||
Protein Name |
Probable histone chaperone asf-1-like protein |
||||||||||||||||||||||||||||||
Protein Synonyms/Alias |
Anti-silencing function protein 1-like; |
||||||||||||||||||||||||||||||
Organism |
Caenorhabditis elegans |
||||||||||||||||||||||||||||||
NCBI Taxonomy ID |
6239 |
||||||||||||||||||||||||||||||
Chromosome Location |
|
||||||||||||||||||||||||||||||
Function in Stage |
|||||||||||||||||||||||||||||||
Function in Cell Type |
|||||||||||||||||||||||||||||||
Description |
Temporarily unavailable |
||||||||||||||||||||||||||||||
The information of related literatures |
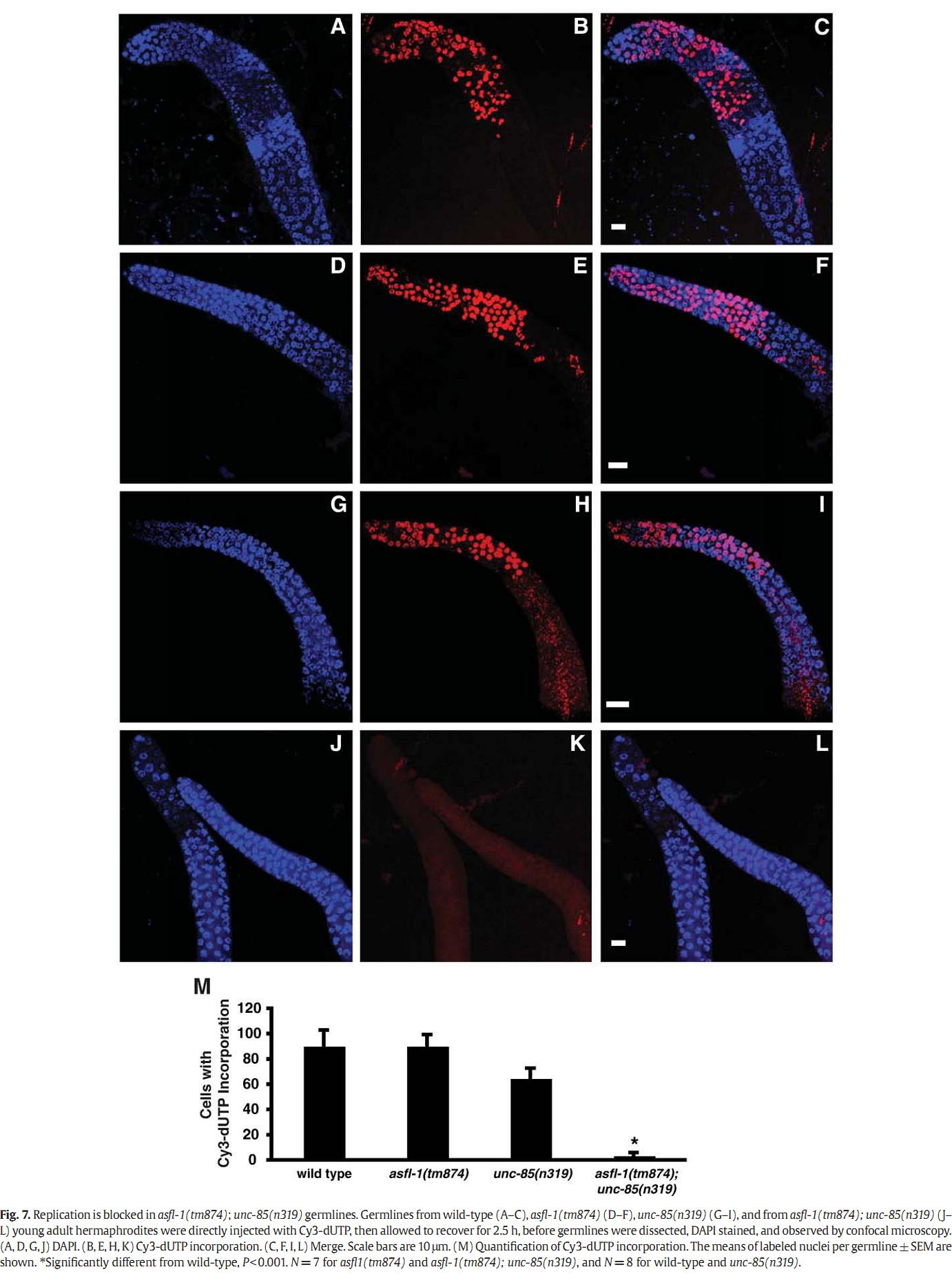
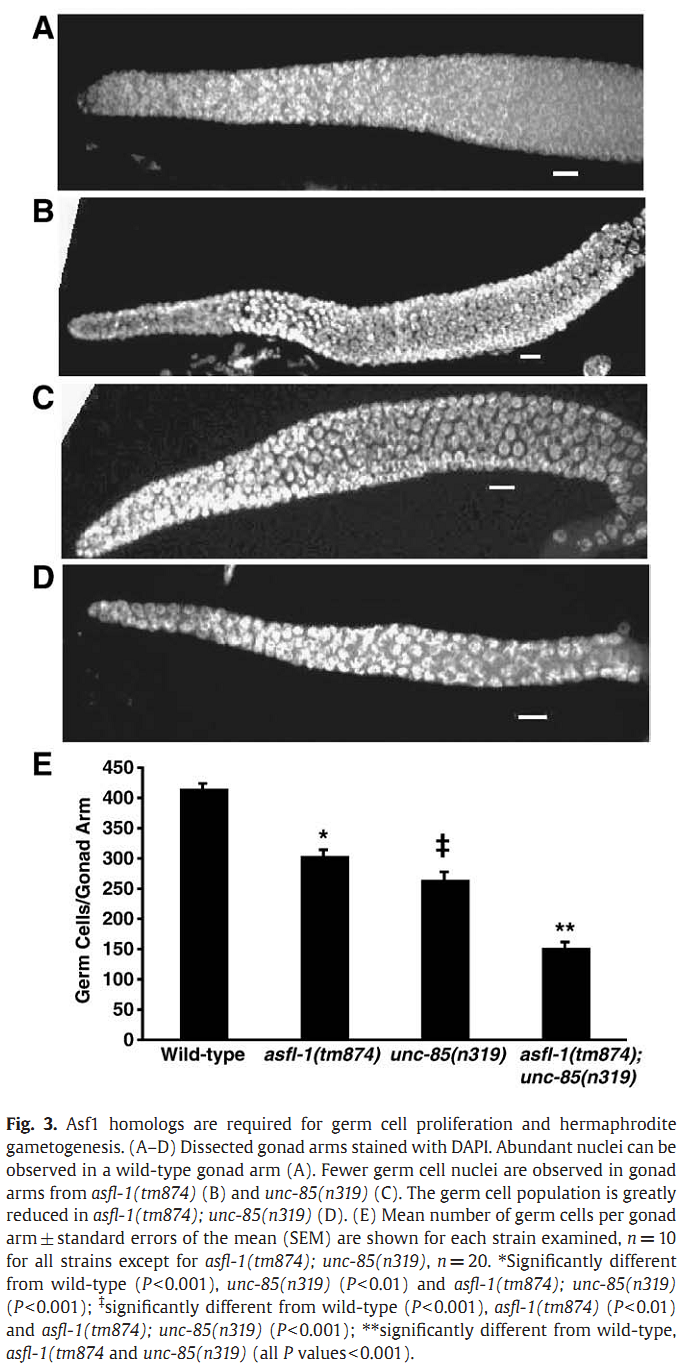
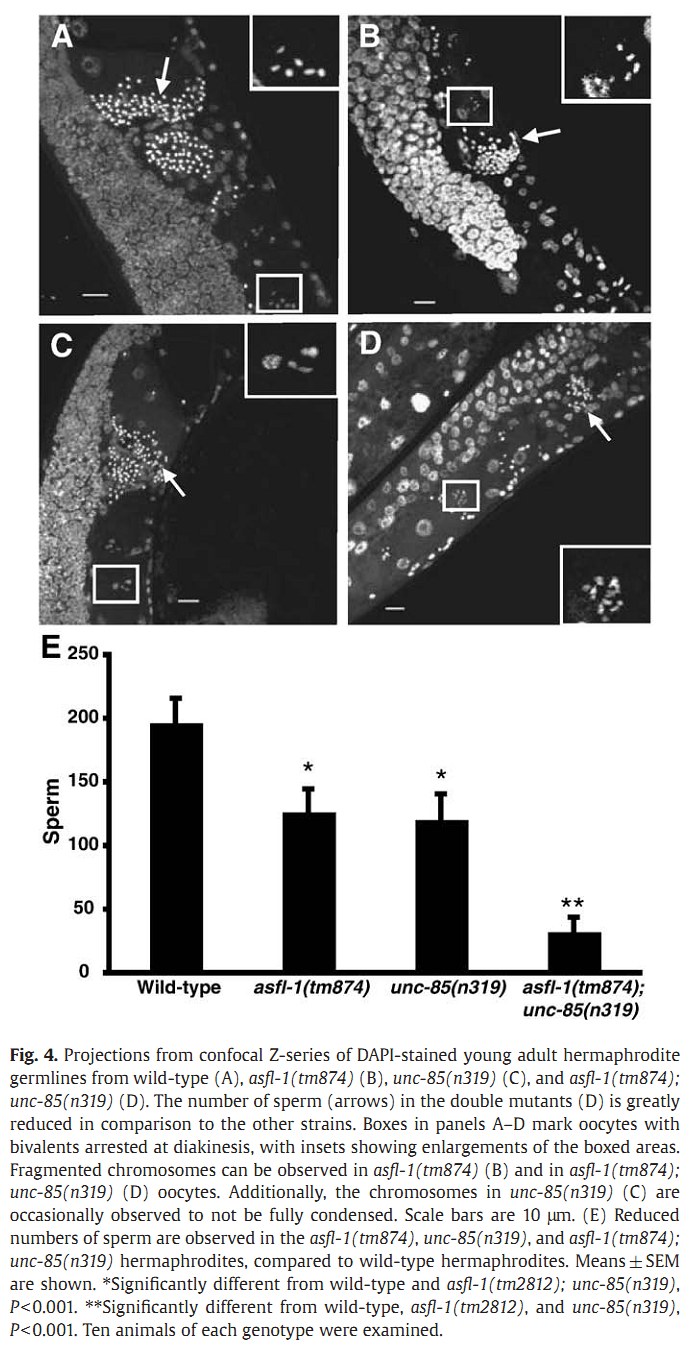
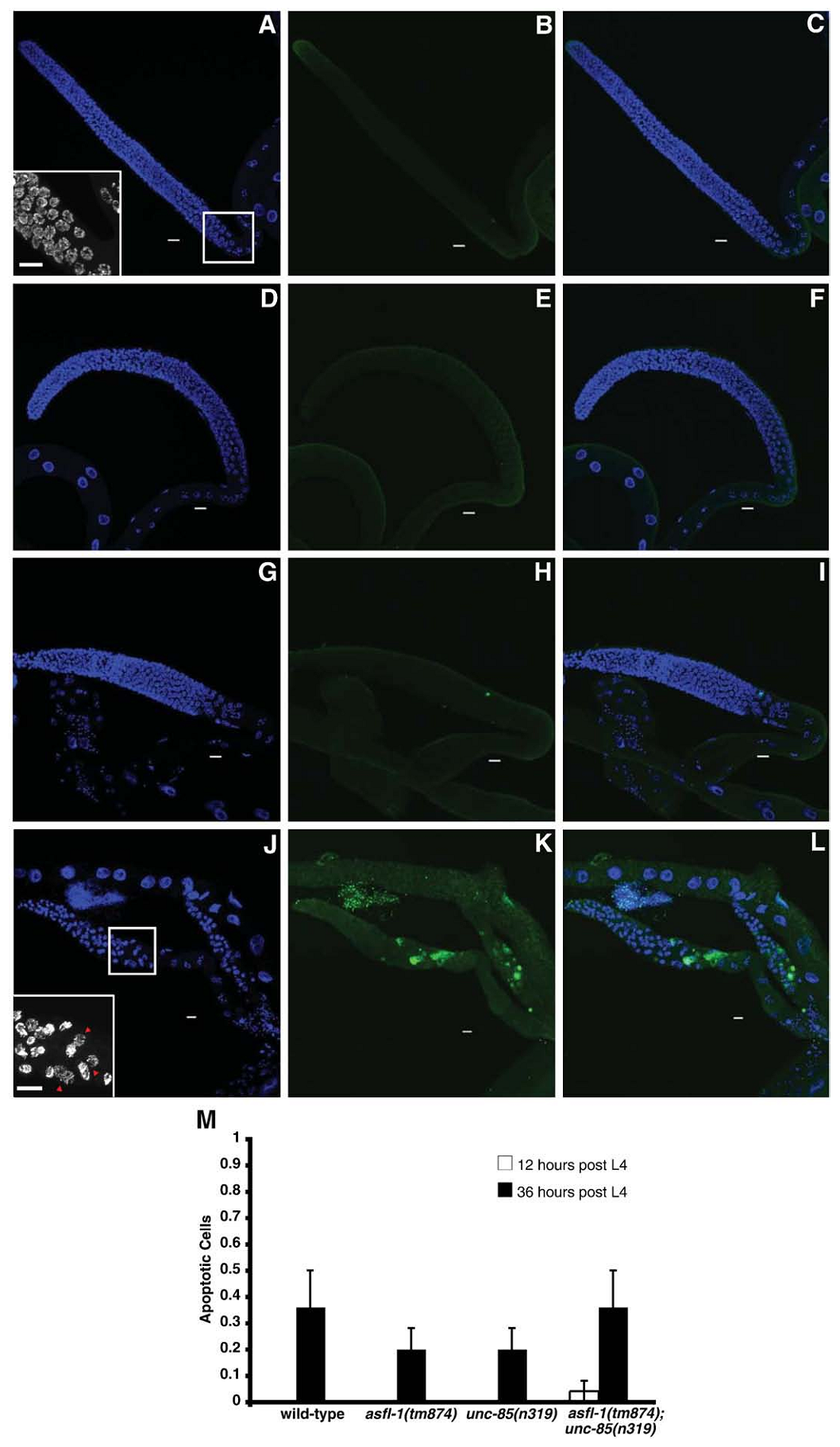
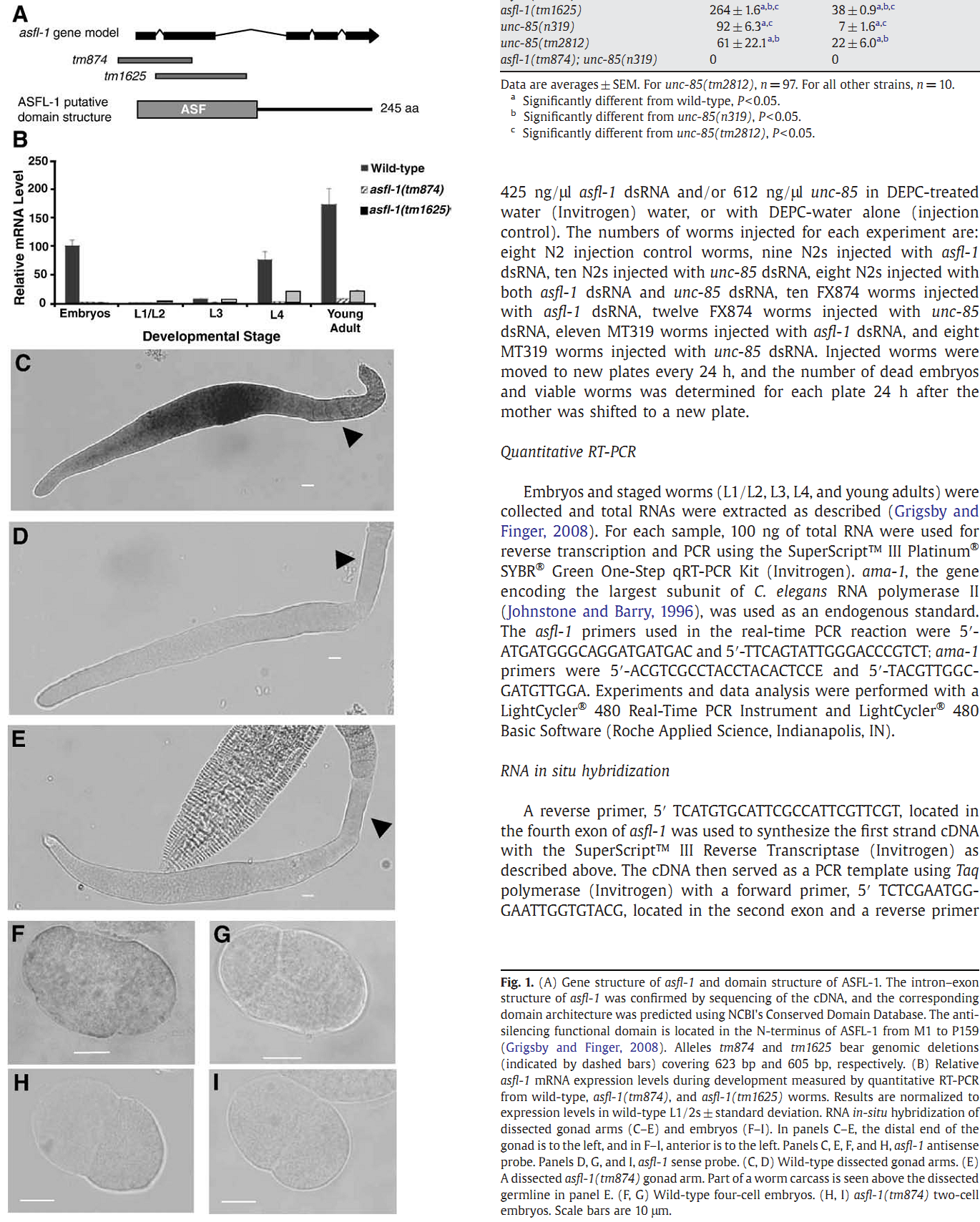
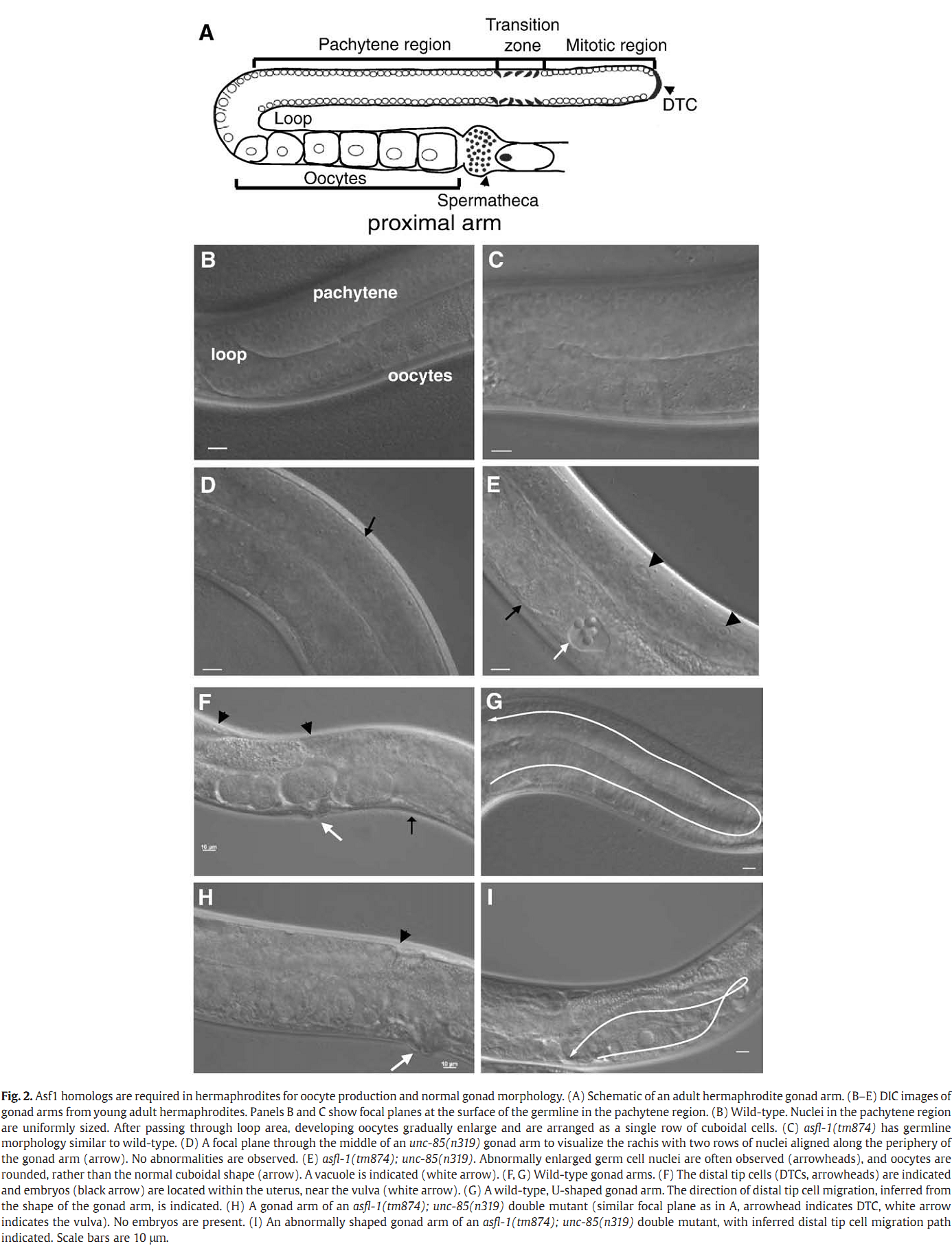
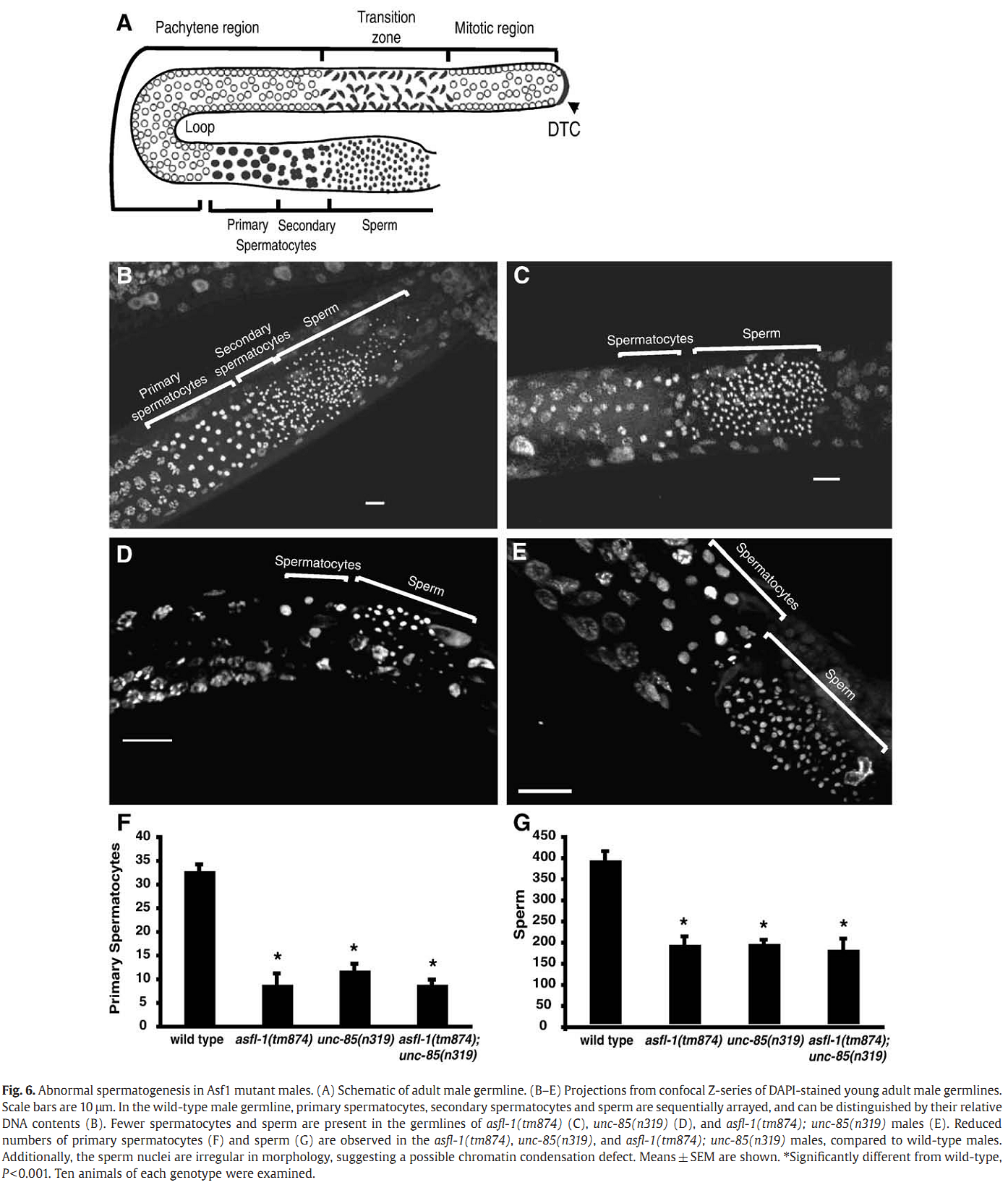
1. I. F. Grigsby, E. M. Rutledge, C. A. Morton and F. P. Finger (2009) Functional redundancy of two C. elegans homologs of the histone chaperone Asf1 in germline DNA replication. Dev Biol 329(1): 64-79. Abstract Eukaryotic genomes contain either one or two genes encoding homologs of the highly conserved histone chaperone Asf1, however, little is known of their in vivo roles in animal development. UNC-85 is one of the two Caenorhabditis elegans Asf1 homologs and functions in post-embryonic replication in neuroblasts. Although UNC-85 is broadly expressed in replicating cells, the specificity of the mutant phenotype suggested possible redundancy with the second C. elegans Asf1 homolog, ASFL-1. The asfl-1 mRNA is expressed in the meiotic region of the germline, and mutants in either Asf1 genes have reduced brood sizes and low penetrance defects in gametogenesis. The asfl-1, unc-85 double mutants are sterile, displaying defects in oogenesis and spermatogenesis, and analysis of DNA synthesis revealed that DNA replication in the germline is blocked. Analysis of somatic phenotypes previously observed in unc-85 mutants revealed that they are neither observed in asfl-1 mutants, nor enhanced in the double mutants, with the exception of enhanced male tail abnormalities in the double mutants. These results suggest that the two Asf1 homologs have partially overlapping functions in the germline, while UNC-85 is primarily responsible for several Asf1 functions in somatic cells, and is more generally involved in replication throughout development. PMID: [19233156] Back to Top |
||||||||||||||||||||||||||||||
Figures for illustrating the function of this protein/gene |
|
||||||||||||||||||||||||||||||
Function |
Histone chaperone that facilitates histone depositionand histone exchange and removal during nucleosome assembly anddisassembly (By similarity). Back to Top |
||||||||||||||||||||||||||||||
Subcellular Location |
Nucleus (By similarity). |
||||||||||||||||||||||||||||||
Tissue Specificity |
|||||||||||||||||||||||||||||||
Gene Ontology |
|
||||||||||||||||||||||||||||||
Interpro |
|||||||||||||||||||||||||||||||
Pfam |
|||||||||||||||||||||||||||||||
SMART |
|||||||||||||||||||||||||||||||
PROSITE |
|||||||||||||||||||||||||||||||
PRINTS |
|||||||||||||||||||||||||||||||
Created Date |
18-Oct-2012 |
||||||||||||||||||||||||||||||
Record Type |
Experiment identified |
||||||||||||||||||||||||||||||
Protein sequence Annotation |
|||||||||||||||||||||||||||||||
Nucleotide Sequence |
Length: 15509 bp Go to nucleotide: FASTA |
||||||||||||||||||||||||||||||
Protein Sequence |
Length: 245 bp Go to amino acid: FASTA |
||||||||||||||||||||||||||||||
The verified Protein-Protein interaction information |
| ||||||||||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
||||||||||||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
||||||||||||||||||||||||||||||
Comments |
|||||||||||||||||||||||||||||||