| Tag | Content | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00000055 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
UniProt Accession |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Theoretical PI |
9.71
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Weight |
79415 Da
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genbank Nucleotide ID |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genbank Protein ID |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Name |
zyg-1 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Synonyms/Alias |
ORFNames=F59E12.2 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Name |
Probable serine/threonine-protein kinase zyg-1 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Synonyms/Alias |
EC=2.7.11.1 Zygote defective protein 1; |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Organism |
Caenorhabditis elegans |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
NCBI Taxonomy ID |
6239 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Chromosome Location |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function in Stage |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function in Cell Type |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Description |
Temporarily unavailable |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The information of related literatures |
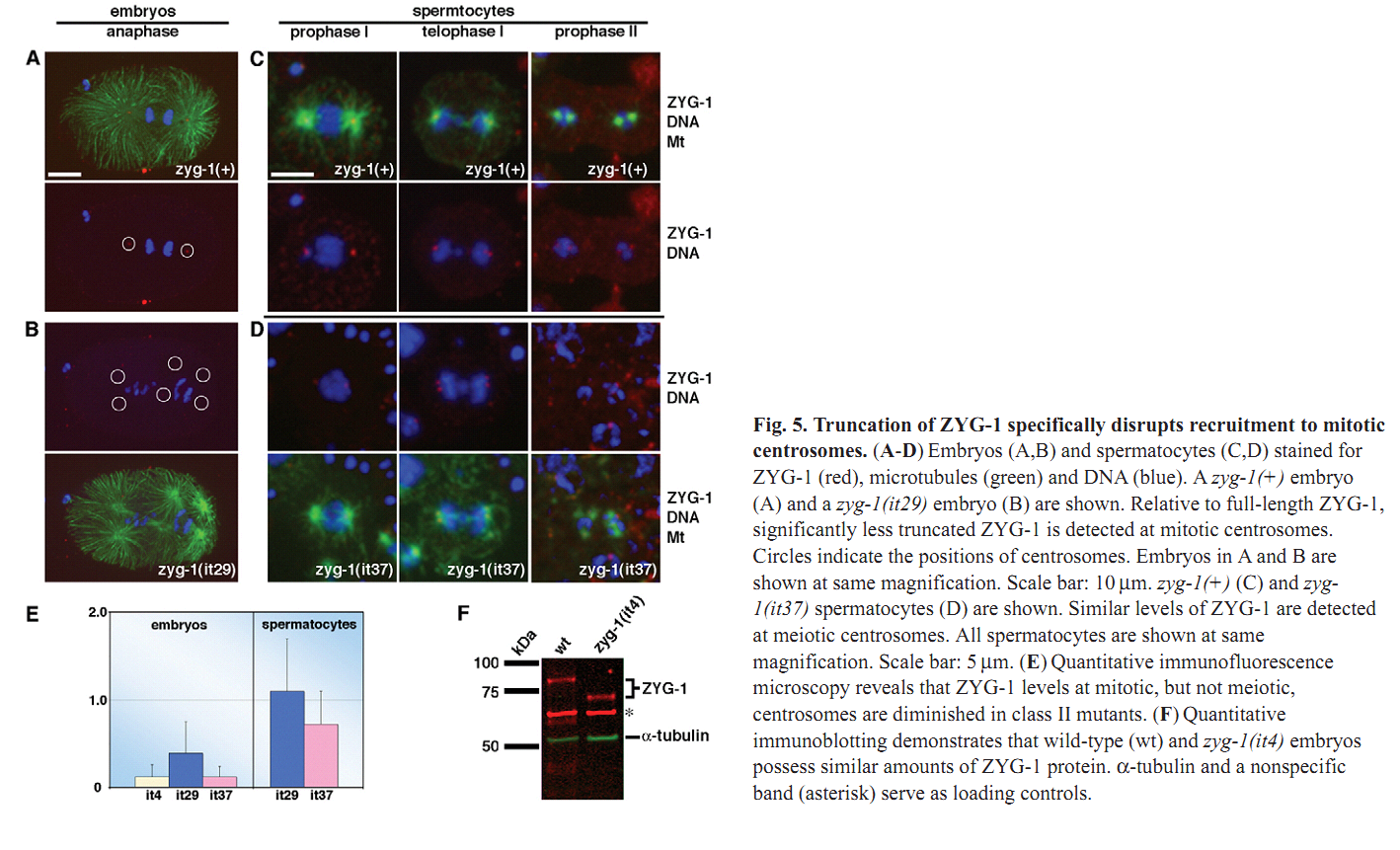
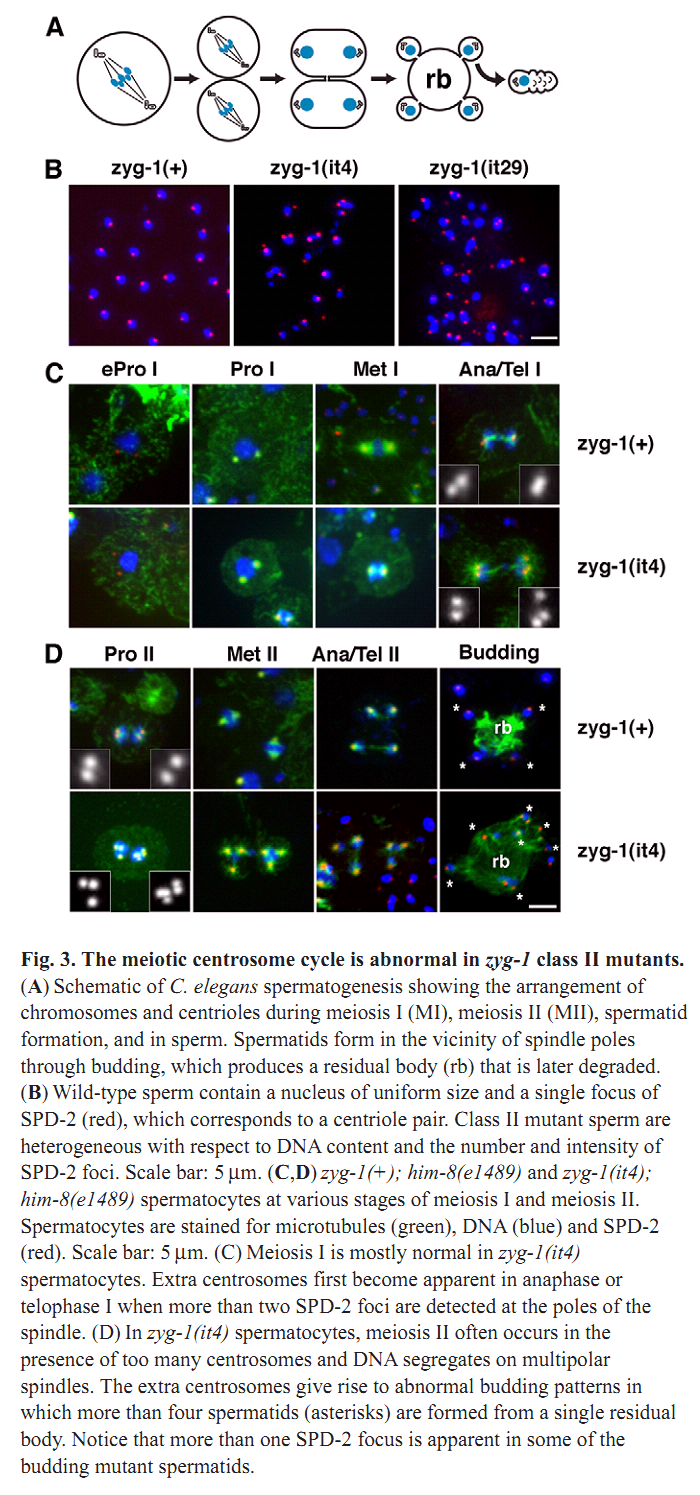
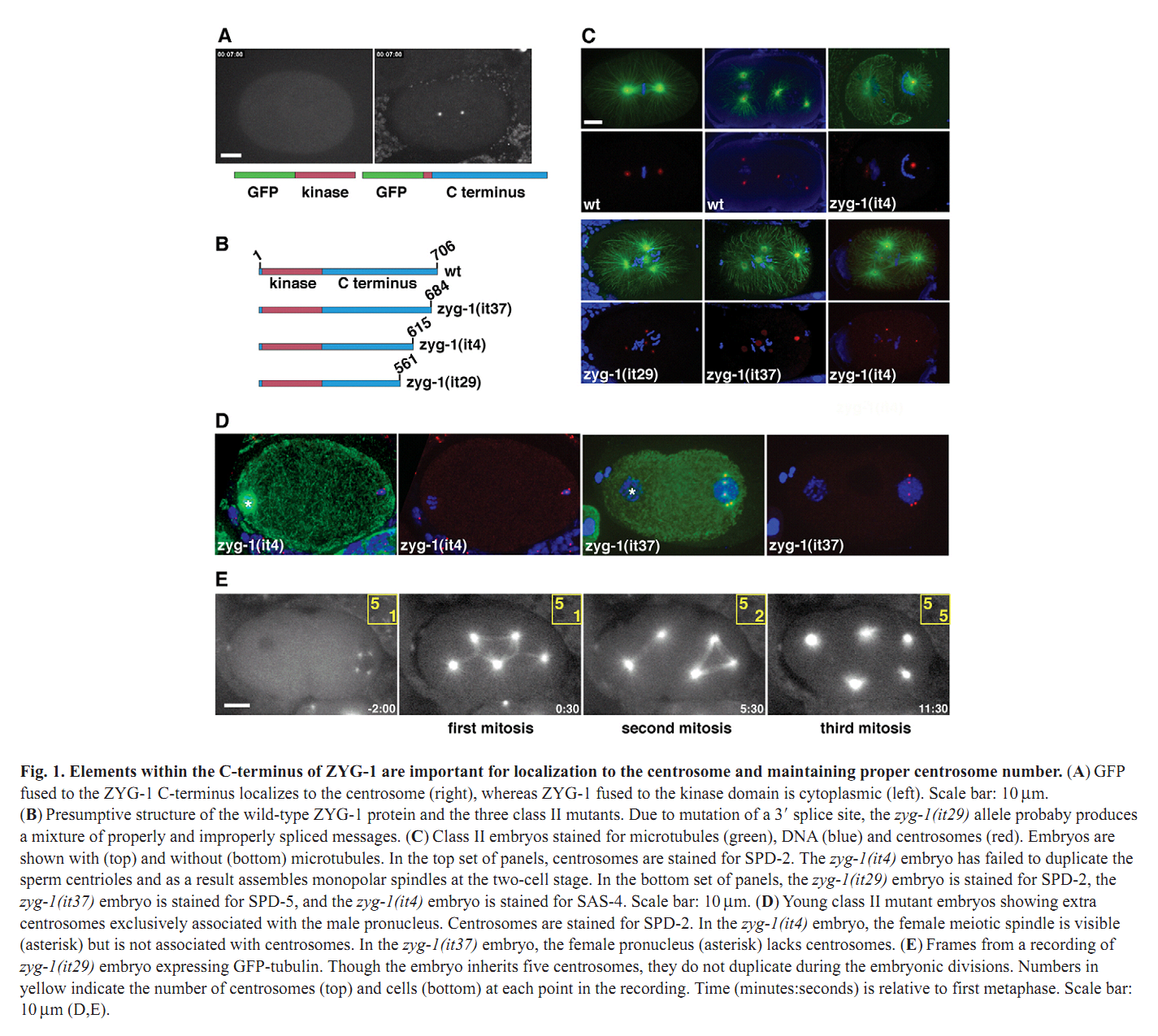
1. N. Peters, D. E. Perez, M. H. Song, Y. Liu, T. Muller-Reichert, C. Caron, K. J. Kemphues and K. F. O'Connell (2010) Control of mitotic and meiotic centriole duplication by the Plk4-related kinase ZYG-1. J Cell Sci 123(Pt 5): 795-805. Abstract Centriole duplication is of crucial importance during both mitotic and male meiotic divisions, but it is currently not known whether this process is regulated differently during the two modes of division. In Caenorhabditis elegans, the kinase ZYG-1 plays an essential role in both mitotic and meiotic centriole duplication. We have found that the C-terminus of ZYG-1 is necessary and sufficient for targeting to centrosomes and is important for differentiating mitotic and meiotic centriole duplication. Small truncations of the C-terminus dramatically lower the level of ZYG-1 at mitotic centrosomes but have little effect on the level of ZYG-1 at meiotic centrosomes. Interestingly, truncation of ZYG-1 blocks centrosome duplication in the mitotic cycle but leads to centrosome amplification in the meiotic cycle. Meiotic centriole amplification appears to result from the overduplication of centrioles during meiosis I and leads to the formation of multipolar meiosis II spindles. The extra centrioles also disrupt spermatogenesis by inducing the formation of supernumerary fertilization-competent spermatids that contain abnormal numbers of chromosomes and centrioles. Our data reveal differences in the regulation of mitotic and meiotic centrosome duplication, particularly with regard to ZYG-1 activity, and reveal an important role for centrosomes in spermatid formation. PMID: [20144993] Back to Top |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figures for illustrating the function of this protein/gene |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function |
Protein kinase that plays a central role in centrosomeduplication. Paternal copy is required to regulate synthesis ofdaughter centrioles prior to fertilization. Maternal copyregulates centrosome duplication during later cell cycles.Functions upstream of sas-5 and sas-6, and is required for theirlocalization to the centrosome. Back to Top |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subcellular Location |
Cytoplasm, cytoskeleton, centrosome.Cytoplasm, cytoskeleton, centrosome, centriole. Note=Component ofthe centrosome. Present at centrioles only immediately beforetheir duplication. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tissue Specificity |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Ontology |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Interpro |
IPR011009; Kinase-like_dom. IPR000719; Prot_kinase_cat_dom. IPR017441; Protein_kinase_ATP_BS. IPR008266; Tyr_kinase_AS. Back to Top |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Pfam |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
SMART |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PROSITE |
PS00107; PROTEIN_KINASE_ATP; 1. PS50011; PROTEIN_KINASE_DOM; 1. PS00109; PROTEIN_KINASE_TYR; 1. Back to Top |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PRINTS |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Created Date |
18-Oct-2012 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Record Type |
Experiment identified |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein sequence Annotation |
CHAIN 1 706 Probable serine/threonine-protein kinase zyg-1. /FTId=PRO_0000086852. DOMAIN 13 249 Protein kinase. NP_BIND 19 27 ATP (By similarity). COMPBIAS 262 381 Arg-rich. ACT_SITE 131 131 Proton acceptor (By similarity). BINDING 41 41 ATP (By similarity). MUTAGEN 41 41 K->M: Loss of function. MUTAGEN 371 371 R->Q: In it4; induces an arrest in cell division at all stages of development. MUTAGEN 432 432 P->L: In b1; induces an arrest in cell division at all stages of development. Back to Top |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Nucleotide Sequence |
Length: 2364 bp Go to nucleotide: FASTA |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Sequence |
Length: 706 bp Go to amino acid: FASTA |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The verified Protein-Protein interaction information |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Comments |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||