| Tag | Content | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00000123 |
||||||||||||||||||||||||
UniProt Accession |
|||||||||||||||||||||||||
Theoretical PI |
UNDEFINED
|
||||||||||||||||||||||||
Molecular Weight |
44342 Da
|
||||||||||||||||||||||||
Genbank Nucleotide ID |
|||||||||||||||||||||||||
Genbank Protein ID |
|||||||||||||||||||||||||
Gene Name |
Dhod |
||||||||||||||||||||||||
Gene Synonyms/Alias |
ORFNames=CG9741 |
||||||||||||||||||||||||
Protein Name |
Dihydroorotate dehydrogenase (quinone), mitochondrial |
||||||||||||||||||||||||
Protein Synonyms/Alias |
DHOdehaseDihydroorotate oxidaseEC=1.3.5.2Flags: Precursor |
||||||||||||||||||||||||
Organism |
Drosophila melanogaster (Fruit fly) |
||||||||||||||||||||||||
NCBI Taxonomy ID |
7227 |
||||||||||||||||||||||||
Chromosome Location |
|
||||||||||||||||||||||||
Function in Stage |
|||||||||||||||||||||||||
Function in Cell Type |
|||||||||||||||||||||||||
Description |
Temporarily unavailable |
||||||||||||||||||||||||
The information of related literatures |
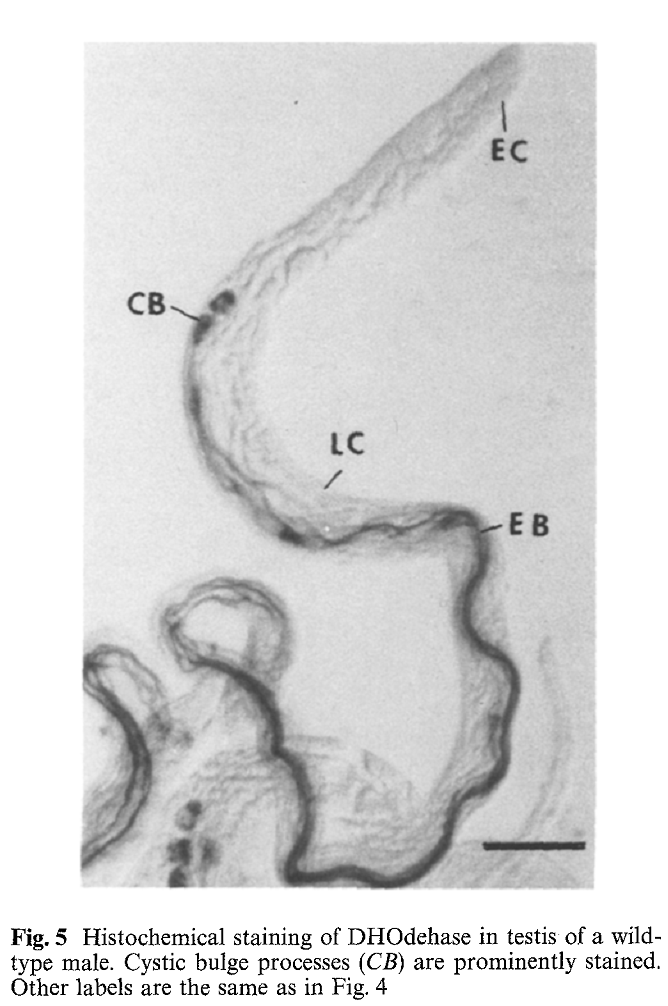
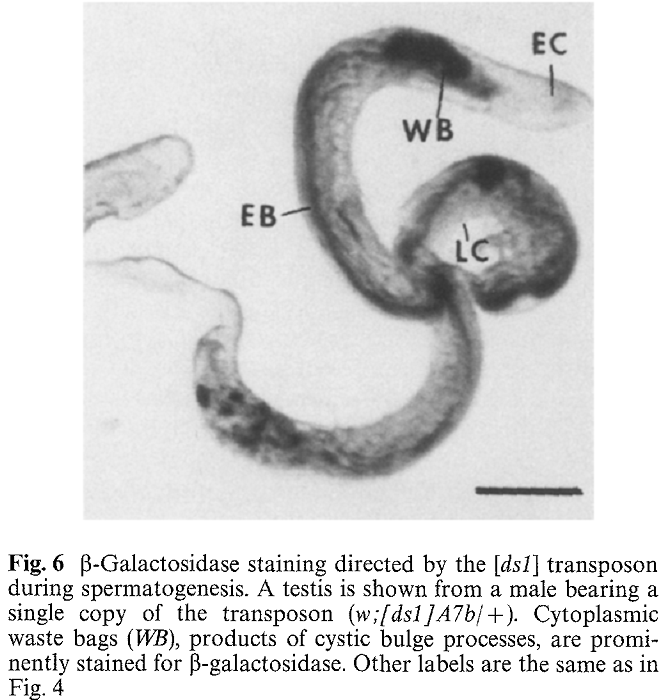
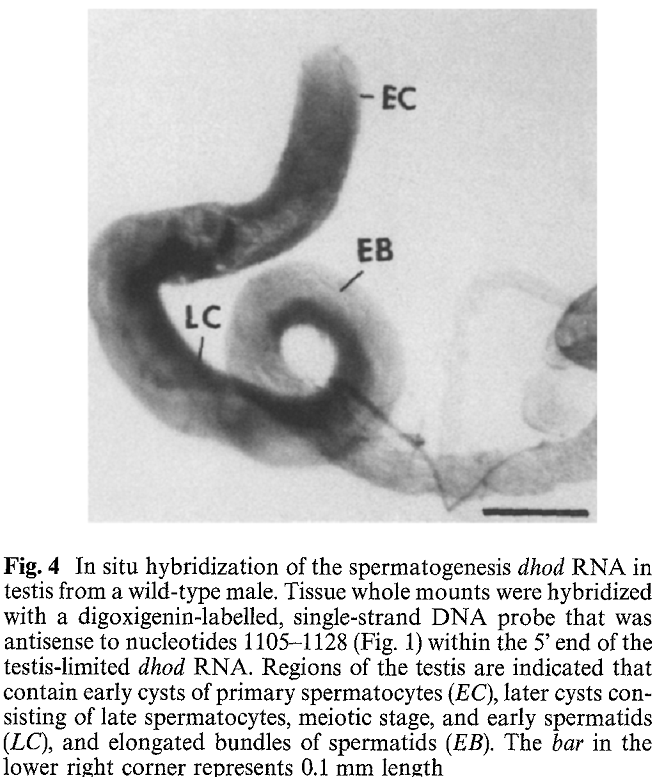
1. J. Yang, L. Porter and J. Rawls (1995) Expression of the dihydroorotate dehydrogenase gene, dhod, during spermatogenesis in Drosophila melanogaster. Mol Gen Genet 246(3): 334-41. Abstract The dhod gene encodes dihydroorotate dehydrogenase (DHOdehase), which catalyzes the fourth step of de novo pyrimidine biosynthesis. In addition to the common 1.5 kb dhod RNA expressed by embryos and females, adult males produce a group of slightly longer RNAs. Evidence is presented that the latter RNAs arise through transcription initiation at sites upstream from that of the common RNA and expression of these male-specific RNAs is limited to spermatogenesis. In situ hybridization analysis shows that these RNAs accumulate during spermatocyte growth and persist through meiosis and early spermatid differentiation. In contrast, DHOdehase activity is virtually absent in spermatocytes, meiotic cells, and in early spermatid cysts, then it becomes highly abundant in elongated spermatid cysts and disappears in late spermatogenesis. Thus, testis-limited expression of dhod conforms to a model proposed for other genes that function during spermiogenesis PMID: [7854318] Back to Top |
||||||||||||||||||||||||
Figures for illustrating the function of this protein/gene |
|
||||||||||||||||||||||||
Function |
Catalyzes the conversion of dihydroorotate to orotatewith quinone as electron acceptor (By similarity). Back to Top |
||||||||||||||||||||||||
Subcellular Location |
Mitochondrion inner membrane; Single-passmembrane protein (Potential). |
||||||||||||||||||||||||
Tissue Specificity |
|||||||||||||||||||||||||
Gene Ontology |
|
||||||||||||||||||||||||
Interpro |
IPR013785; Aldolase_TIM. IPR012135; Dihydroorotate_DH_1_2. IPR005719; Dihydroorotate_DH_2. IPR001295; Dihydroorotate_DH_CS. Back to Top |
||||||||||||||||||||||||
Pfam |
|||||||||||||||||||||||||
SMART |
|||||||||||||||||||||||||
PROSITE |
|||||||||||||||||||||||||
PRINTS |
|||||||||||||||||||||||||
Created Date |
18-Oct-2012 |
||||||||||||||||||||||||
Record Type |
Experiment identified |
||||||||||||||||||||||||
Protein sequence Annotation |
TRANSIT 1 ? Mitochondrion (Potential). CHAIN ? 405 Dihydroorotate dehydrogenase (quinone), mitochondrial. /FTId=PRO_0000029887. TOPO_DOM ? 20 Mitochondrial matrix (Potential). TRANSMEM 21 39 Helical; (Potential). TOPO_DOM 40 401 Mitochondrial intermembrane (Potential). NP_BIND 103 107 FMN (By similarity). NP_BIND 361 362 FMN (By similarity). REGION 152 156 Substrate binding (By similarity). REGION 215 220 Substrate binding (By similarity). REGION 289 290 Substrate binding (By similarity). ACT_SITE 218 218 Nucleophile (By similarity). BINDING 107 107 Substrate (By similarity). BINDING 127 127 FMN (By similarity). BINDING 184 184 FMN (By similarity). BINDING 215 215 FMN (By similarity). BINDING 215 215 Substrate (By similarity). BINDING 258 258 FMN (By similarity). BINDING 288 288 FMN; via carbonyl oxygen (By similarity). BINDING 311 311 FMN; via amide nitrogen (By similarity). BINDING 340 340 FMN; via amide nitrogen (By similarity). Back to Top |
||||||||||||||||||||||||
Nucleotide Sequence |
Length: 3078 bp Go to nucleotide: FASTA |
||||||||||||||||||||||||
Protein Sequence |
Length: 405 bp Go to amino acid: FASTA |
||||||||||||||||||||||||
The verified Protein-Protein interaction information |
|||||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
||||||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
||||||||||||||||||||||||
Comments |
|||||||||||||||||||||||||