| Tag | Content | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00000126 |
||||||||||||||||||||||||||||||
UniProt Accession |
|||||||||||||||||||||||||||||||
Theoretical PI |
5.43
|
||||||||||||||||||||||||||||||
Molecular Weight |
160133 Da
|
||||||||||||||||||||||||||||||
Genbank Nucleotide ID |
|||||||||||||||||||||||||||||||
Genbank Protein ID |
|||||||||||||||||||||||||||||||
Gene Name |
kel |
||||||||||||||||||||||||||||||
Gene Synonyms/Alias |
ORFNames=CG7210 |
||||||||||||||||||||||||||||||
Protein Name |
Ring canal kelch protein Kelch short protein |
||||||||||||||||||||||||||||||
Protein Synonyms/Alias |
Contains: |
||||||||||||||||||||||||||||||
Organism |
Drosophila melanogaster (Fruit fly) |
||||||||||||||||||||||||||||||
NCBI Taxonomy ID |
7227 |
||||||||||||||||||||||||||||||
Chromosome Location |
|
||||||||||||||||||||||||||||||
Function in Stage |
|||||||||||||||||||||||||||||||
Function in Cell Type |
|||||||||||||||||||||||||||||||
Description |
Temporarily unavailable |
||||||||||||||||||||||||||||||
The information of related literatures |
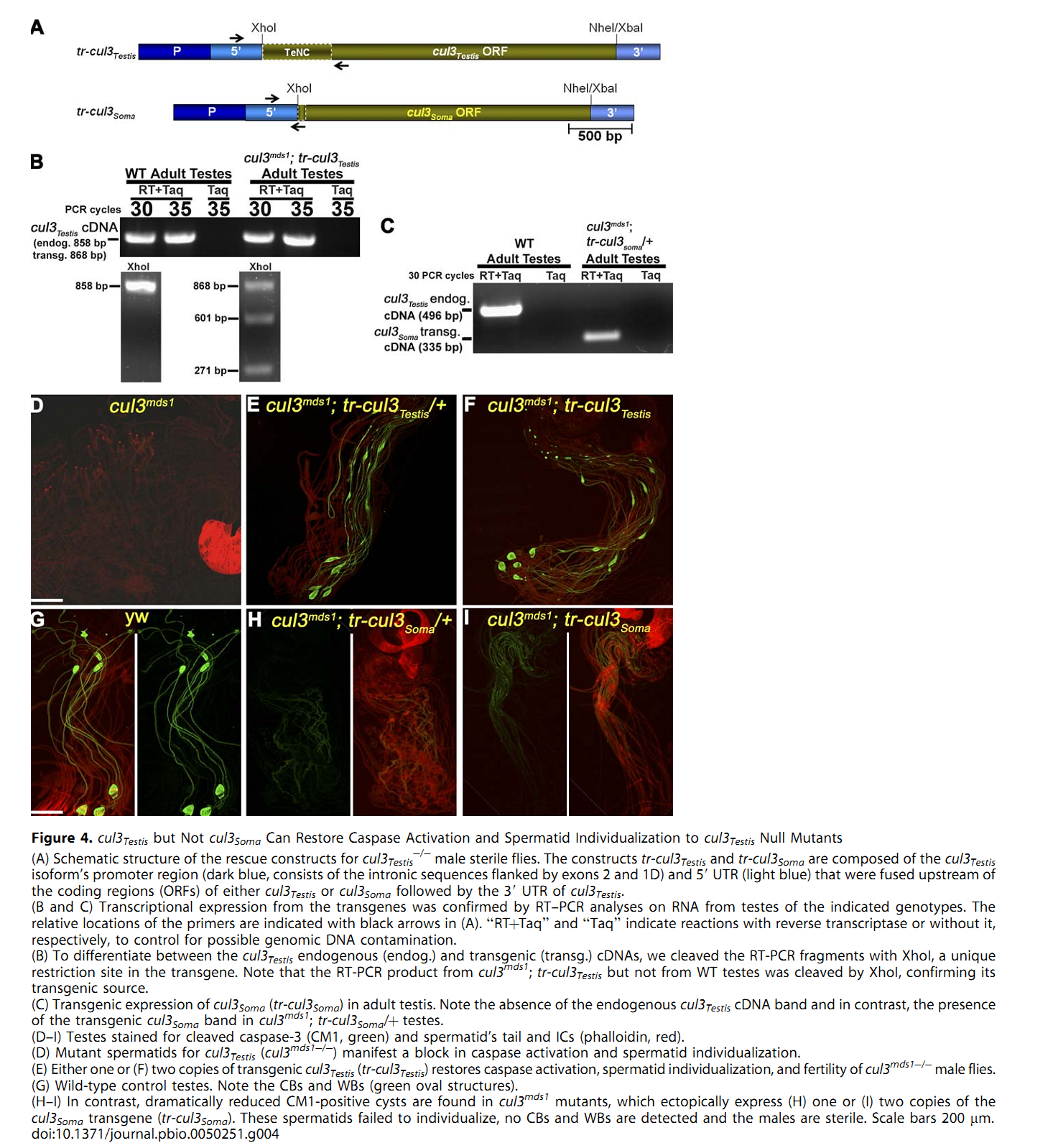
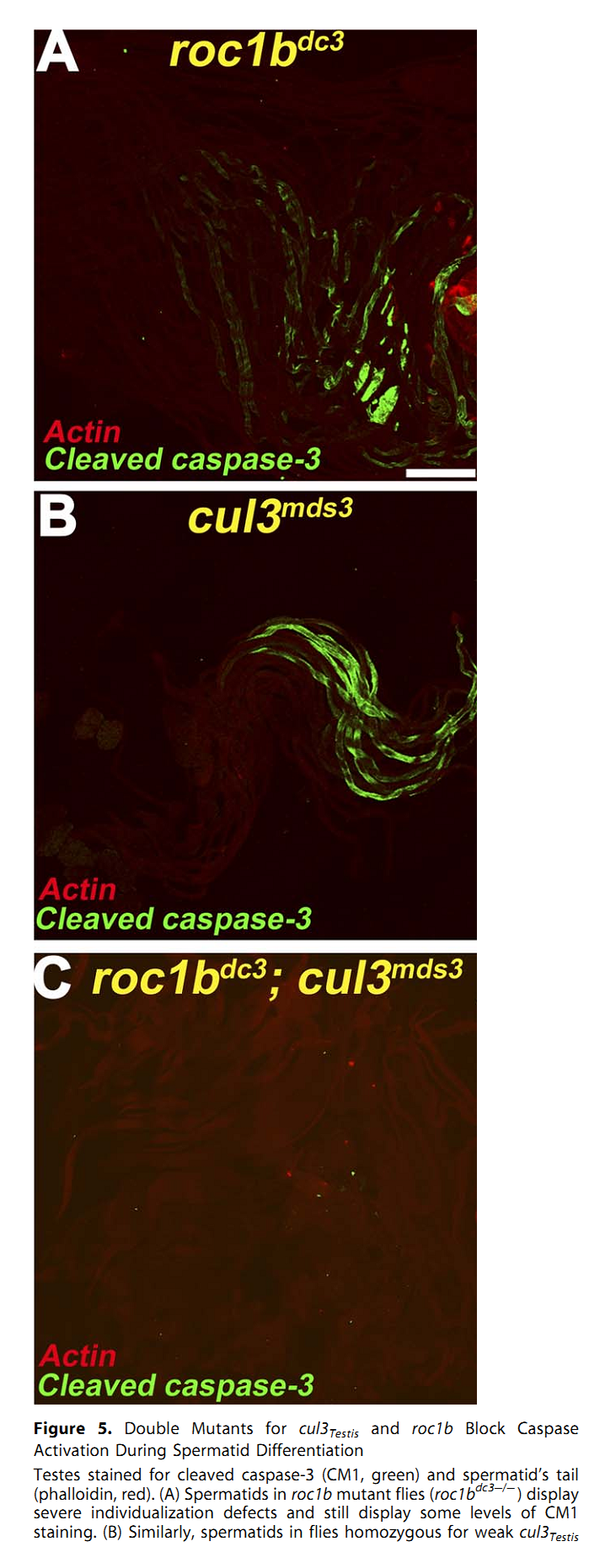
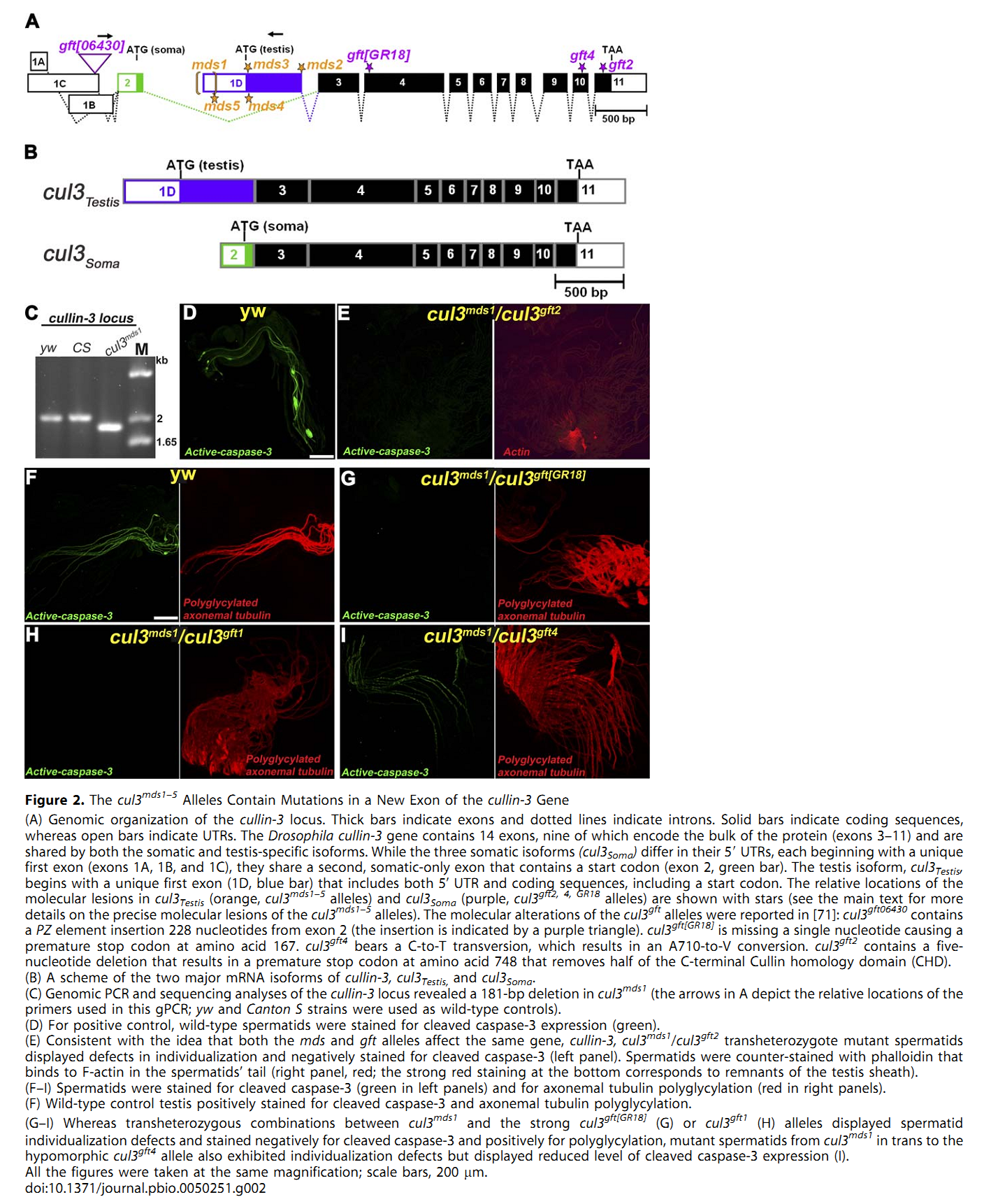
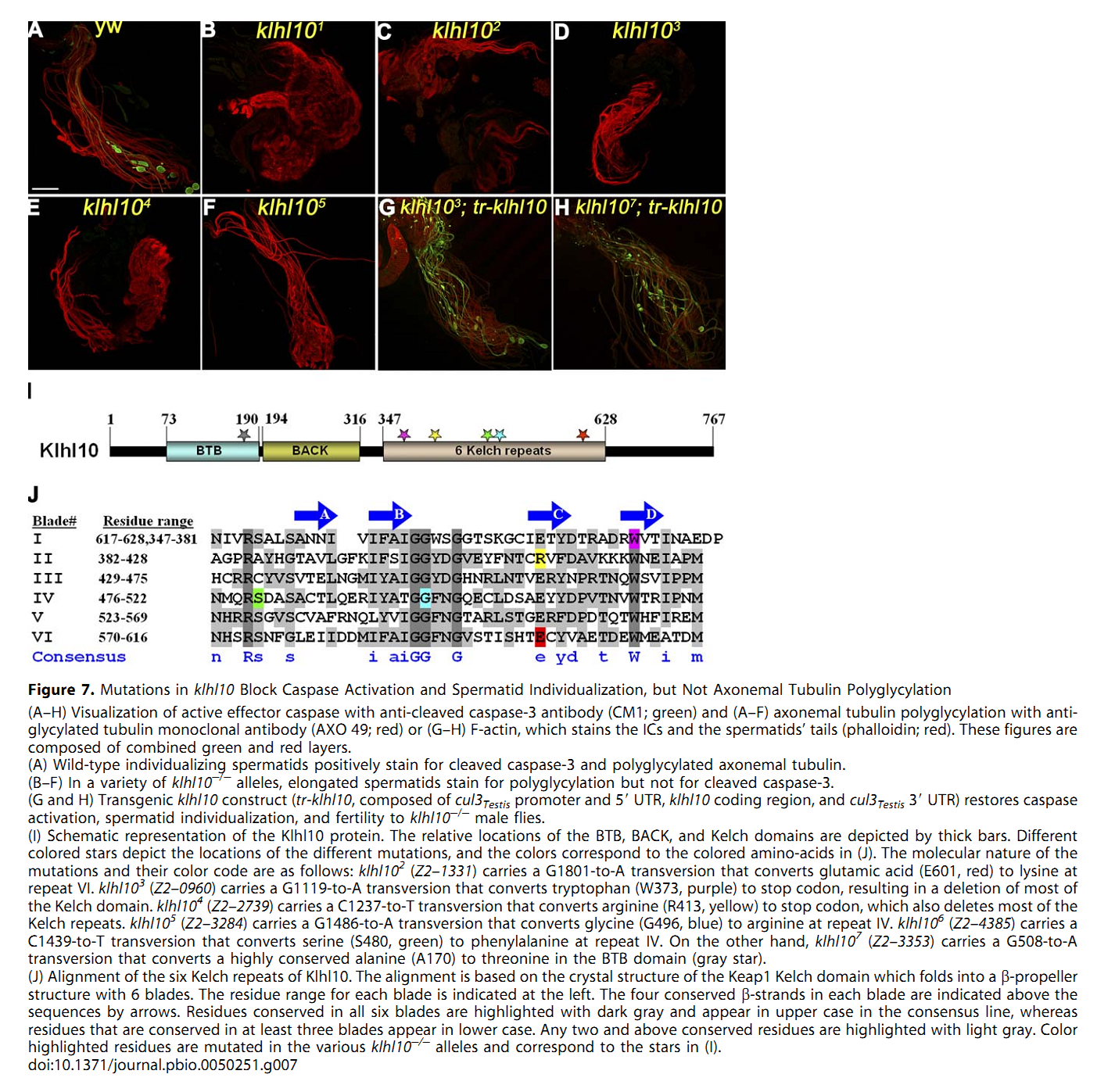
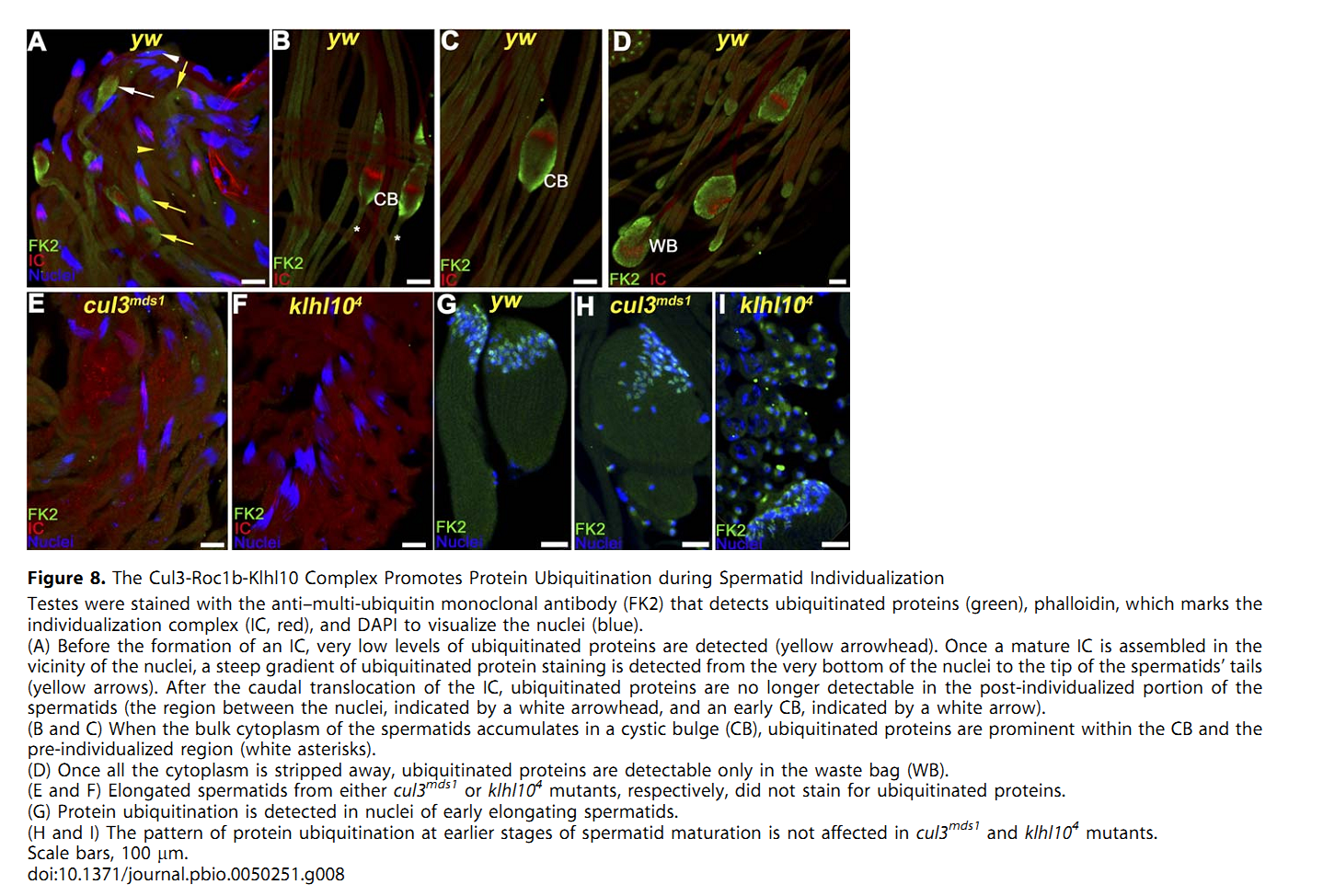
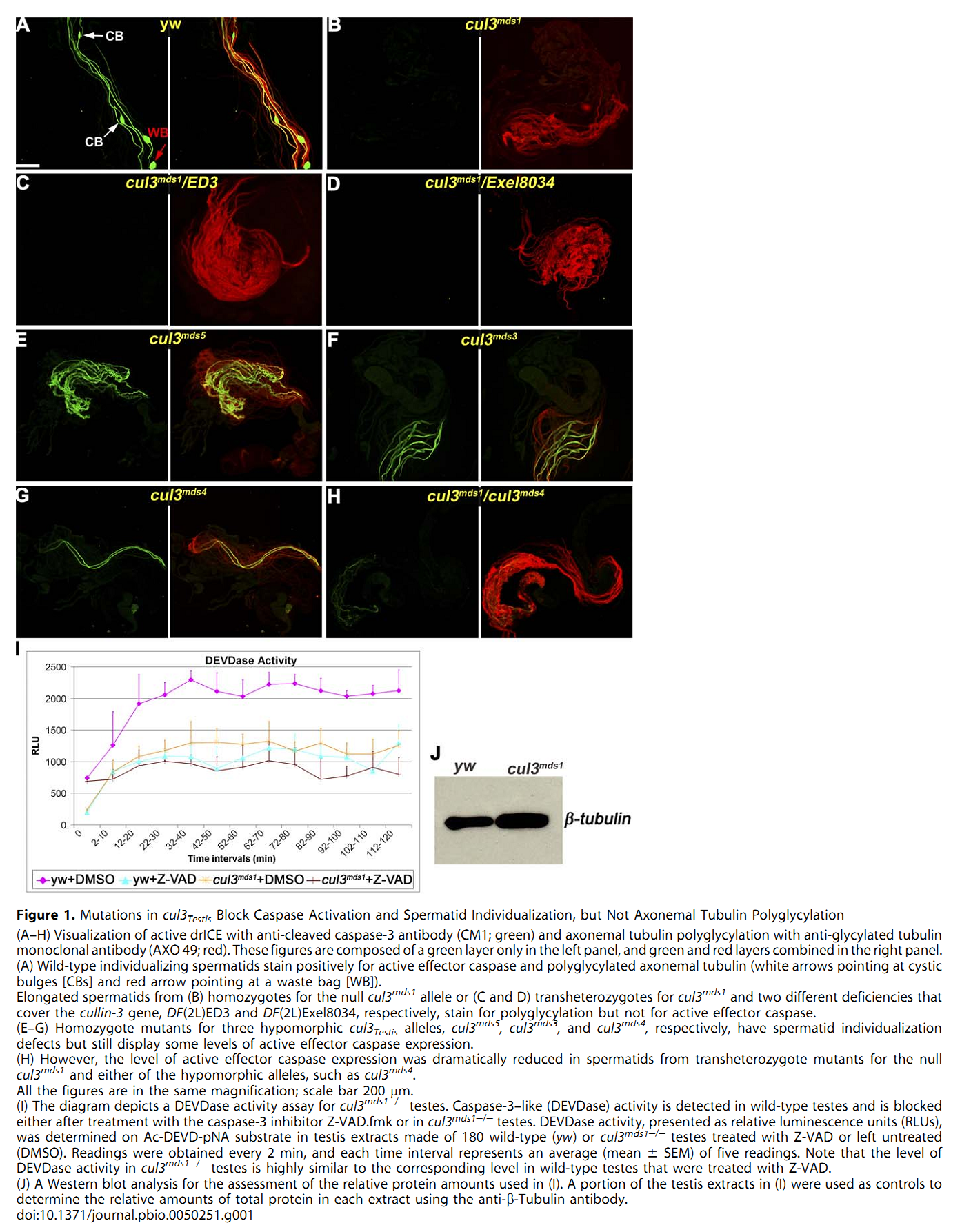
1. E. Arama, M. Bader, G. E. Rieckhof and H. Steller (2007) A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol 5(10): e251. Abstract In both insects and mammals, spermatids eliminate their bulk cytoplasm as they undergo terminal differentiation. In Drosophila, this process of dramatic cellular remodeling requires apoptotic proteins, including caspases. To gain further insight into the regulation of caspases, we screened a large collection of sterile male flies for mutants that block effector caspase activation at the onset of spermatid individualization. Here, we describe the identification and characterization of a testis-specific, Cullin-3-dependent ubiquitin ligase complex that is required for caspase activation in spermatids. Mutations in either a testis-specific isoform of Cullin-3 (Cul3(Testis)), the small RING protein Roc1b, or a Drosophila orthologue of the mammalian BTB-Kelch protein Klhl10 all reduce or eliminate effector caspase activation in spermatids. Importantly, all three genes encode proteins that can physically interact to form a ubiquitin ligase complex. Roc1b binds to the catalytic core of Cullin-3, and Klhl10 binds specifically to a unique testis-specific N-terminal Cullin-3 (TeNC) domain of Cul3(Testis) that is required for activation of effector caspase in spermatids. Finally, the BIR domain region of the giant inhibitor of apoptosis-like protein dBruce is sufficient to bind to Klhl10, which is consistent with the idea that dBruce is a substrate for the Cullin-3-based E3-ligase complex. These findings reveal a novel role of Cullin-based ubiquitin ligases in caspase regulation. PMID: [17880263] Back to Top |
||||||||||||||||||||||||||||||
Figures for illustrating the function of this protein/gene |
|
||||||||||||||||||||||||||||||
Function |
Component of ring canals that regulates the flow ofcytoplasm between cells. May be involved in the regulation ofcytoplasm flow from nurse cells to the oocyte during oogenesis.Binds actin. Back to Top |
||||||||||||||||||||||||||||||
Subcellular Location |
Cytoplasm, cytoskeleton. Note=Inner surfaceof cytoplasmic bridges or ring canals present in egg chambers.Subcortically in imaginal disk epithelia. |
||||||||||||||||||||||||||||||
Tissue Specificity |
Both proteins are expressed in ovaries, maletestis, ovariectomized females, cuticle, salivary gland andimaginal disks. Kelch short protein is the predominant form and isalso expressed in fat bodies. On entry into metamorphosis levelsof full length protein increase in testis and imaginal disks. |
||||||||||||||||||||||||||||||
Gene Ontology |
|
||||||||||||||||||||||||||||||
Interpro |
|||||||||||||||||||||||||||||||
Pfam |
|||||||||||||||||||||||||||||||
SMART |
|||||||||||||||||||||||||||||||
PROSITE |
|||||||||||||||||||||||||||||||
PRINTS |
|||||||||||||||||||||||||||||||
Created Date |
18-Oct-2012 |
||||||||||||||||||||||||||||||
Record Type |
Experiment identified |
||||||||||||||||||||||||||||||
Protein sequence Annotation |
CHAIN 1 1477 Ring canal kelch protein. /FTId=PRO_0000016651. CHAIN 1 689 Kelch short protein. /FTId=PRO_0000016652. DOMAIN 157 223 BTB. REPEAT 404 449 Kelch 1. REPEAT 450 496 Kelch 2. REPEAT 498 543 Kelch 3. REPEAT 545 592 Kelch 4. REPEAT 594 639 Kelch 5. REPEAT 641 687 Kelch 6. COMPBIAS 18 28 Asn-rich. COMPBIAS 29 87 Gln-rich. COMPBIAS 29 36 Poly-Gln. COMPBIAS 78 83 Poly-Gln. NON_STD 690 690 Selenocysteine (Probable). MOD_RES 108 108 Phosphoserine. MOD_RES 111 111 Phosphoserine. CONFLICT 493 493 V -> A (in Ref. 1; AAA53471/AAA53472). CONFLICT 596 596 A -> R (in Ref. 1; AAA53471/AAA53472). CONFLICT 824 824 P -> L (in Ref. 1; AAA53472). CONFLICT 858 858 G -> D (in Ref. 1; AAA53472). CONFLICT 1083 1083 A -> R (in Ref. 1; AAA53472). CONFLICT 1086 1086 A -> G (in Ref. 1; AAA53472). Back to Top |
||||||||||||||||||||||||||||||
Nucleotide Sequence |
Length: 5619 bp Go to nucleotide: FASTA |
||||||||||||||||||||||||||||||
Protein Sequence |
Length: 1477 bp Go to amino acid: FASTA |
||||||||||||||||||||||||||||||
The verified Protein-Protein interaction information |
|||||||||||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
||||||||||||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
||||||||||||||||||||||||||||||
Comments |
|||||||||||||||||||||||||||||||