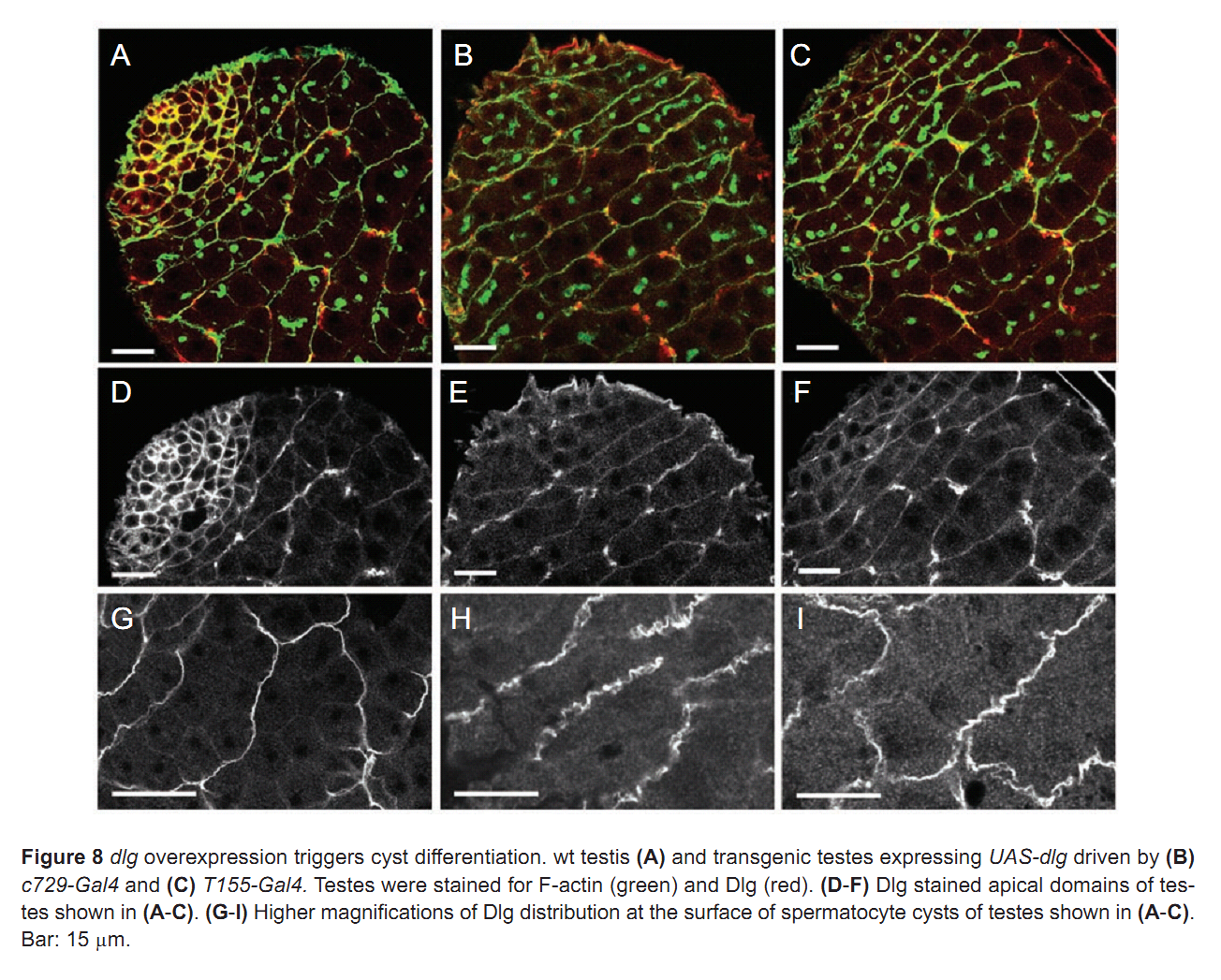
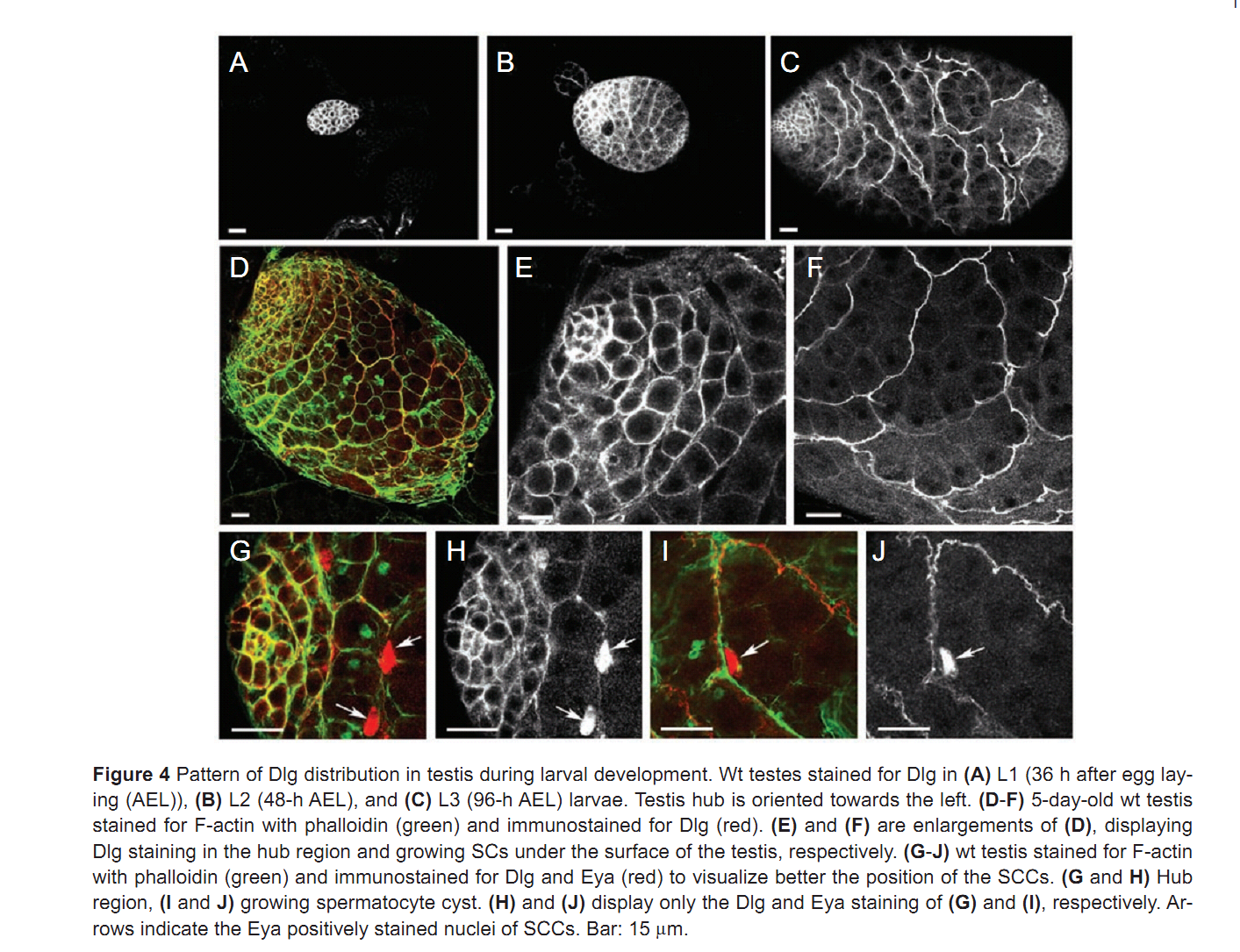
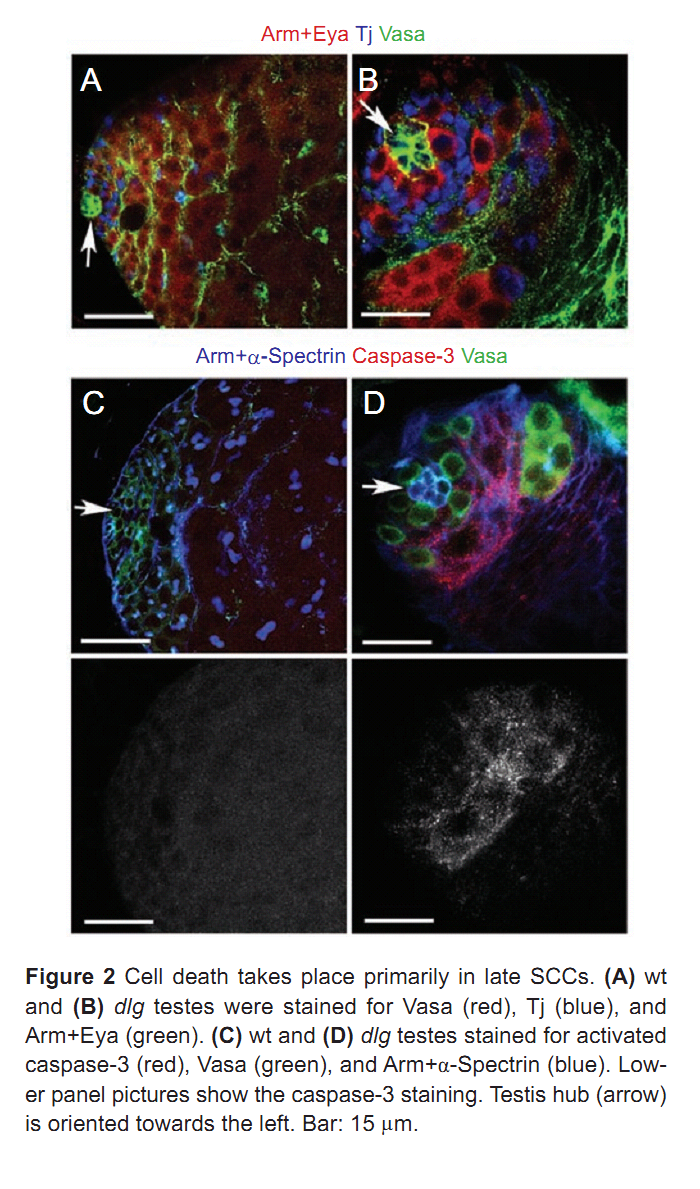
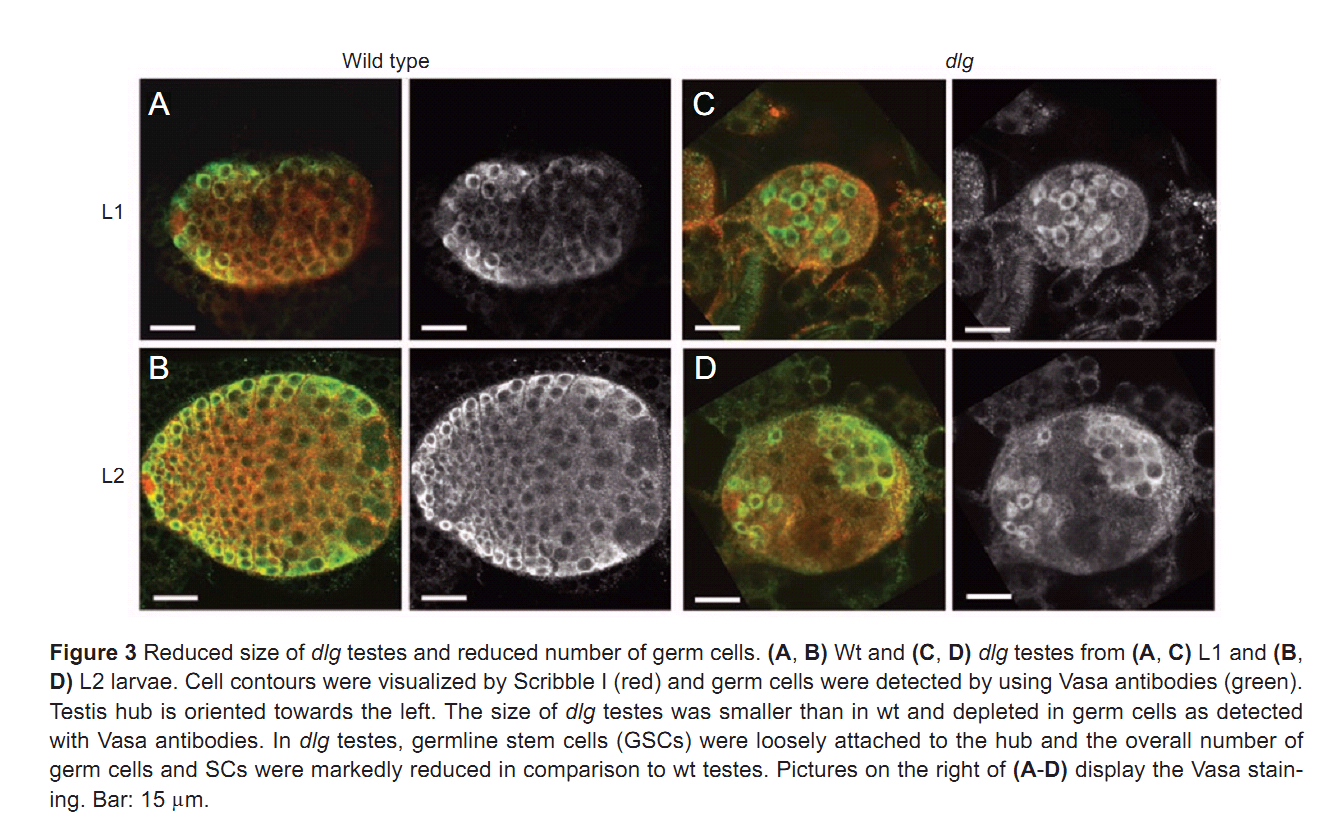
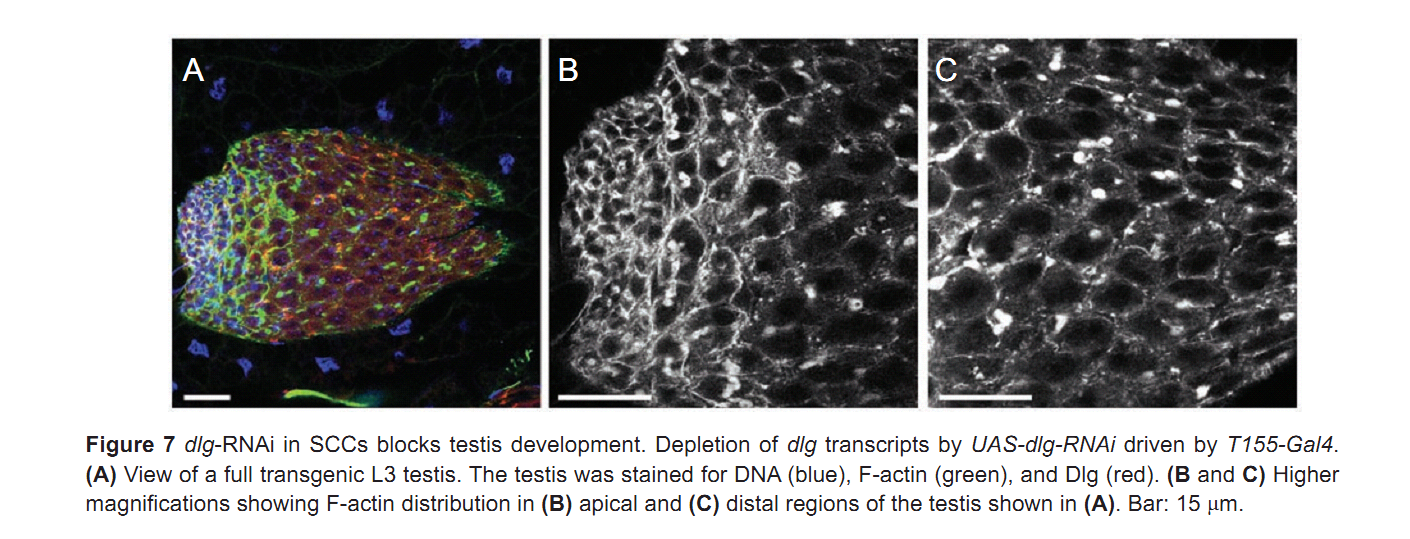
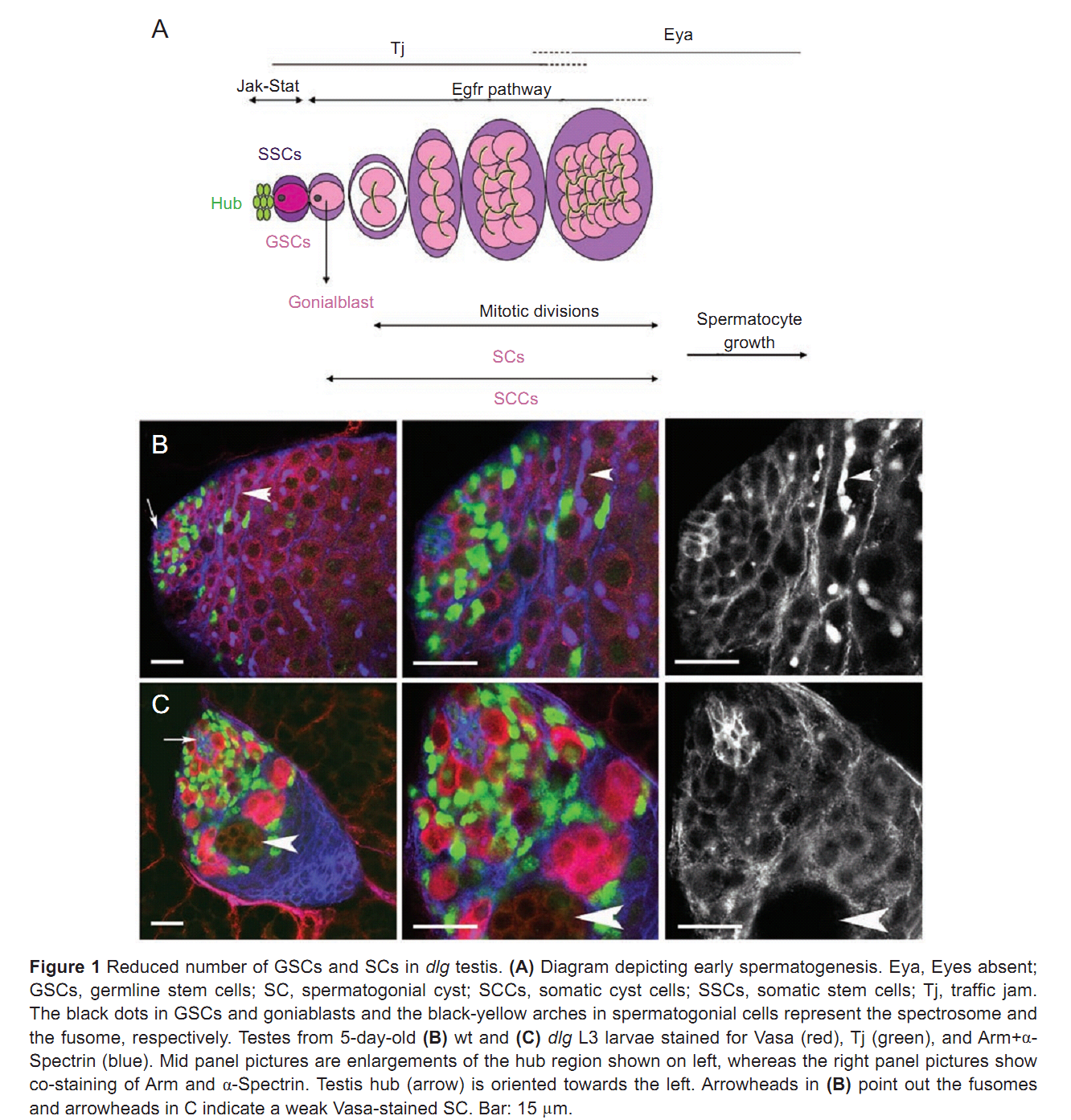
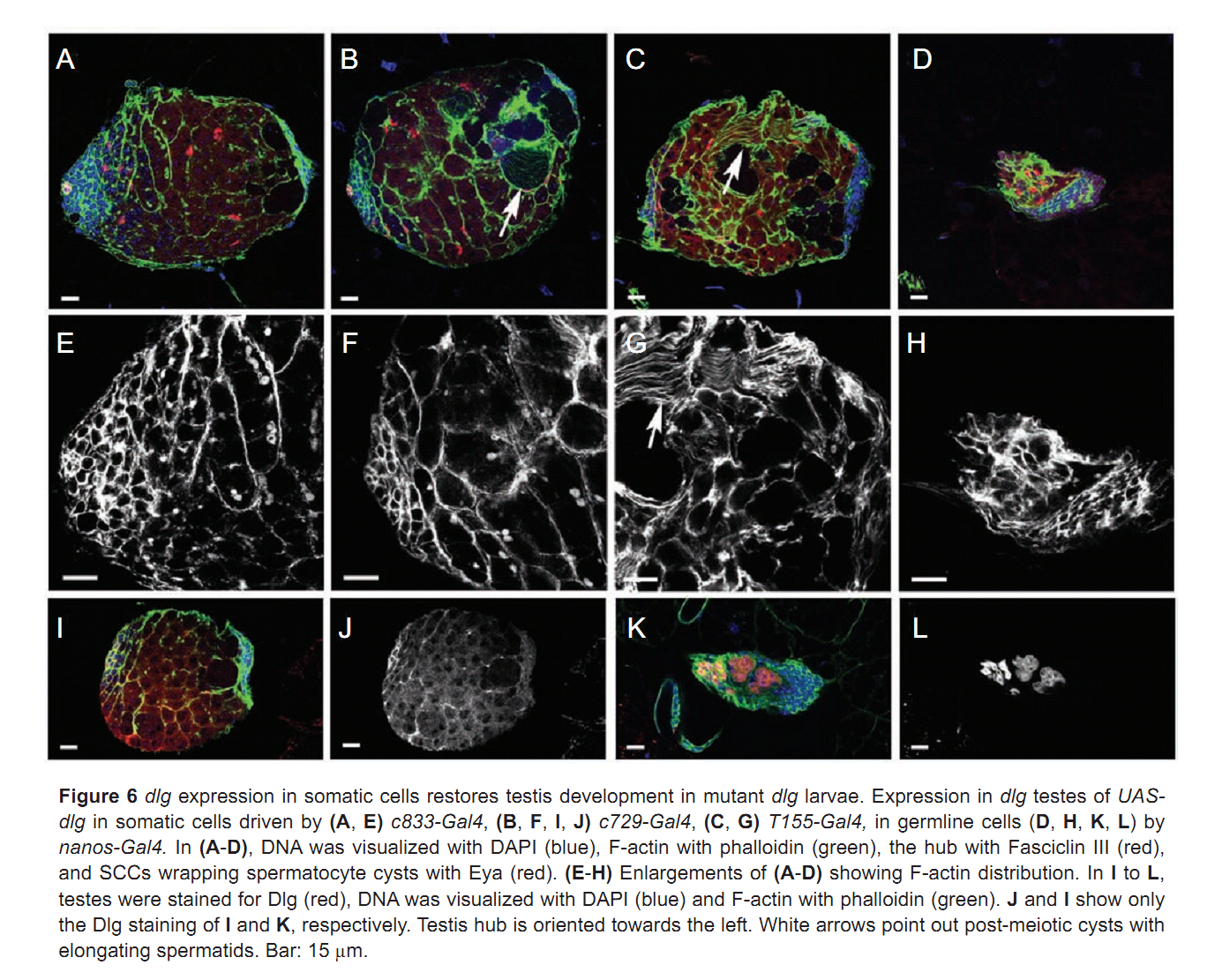
| Tag | Content | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00000128 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
UniProt Accession |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Theoretical PI |
6.31
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Weight |
106673 Da
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genbank Nucleotide ID |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genbank Protein ID |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Name |
dlg1 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Synonyms/Alias |
l(1)dlg1 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Name |
Disks large 1 tumor suppressor protein |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Synonyms/Alias |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Organism |
Drosophila melanogaster (Fruit fly) |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
NCBI Taxonomy ID |
7227 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Chromosome Location |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function in Stage |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function in Cell Type |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Description |
Temporarily unavailable |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The information of related literatures |
1. F. Papagiannouli and B. M. Mechler (2009) discs large regulates somatic cyst cell survival and expansion in Drosophila testis. Cell Res 19(10): 1139-49. Abstract Gonad development requires a coordinated soma-germline interaction that ensures renewal and differentiation of germline and somatic stem cells to ultimately produce mature gametes. The Drosophila tumour suppressor gene discs large (dlg) encodes a septate junction protein functioning during epithelial polarization, asymmetric neuroblast division, and formation of neuromuscular junctions. Here, we report the role of dlg in testis development and its critical function in somatic cyst cells (SCCs). In these cells dlg is primarily required for their survival and expansion, and contributes to spermatocyte cyst differentiation. Cell death primarily occurred in SCCs at the end of spermatogonial amplification at a time when Dlg becomes restricted in wild-type (wt) testes to the distal somatic cells capping the growing spermatocyte cysts. RNAi depletion of dlg transcripts in early SCCs fully prevented testis development, whereas depletion in late SCCs resulted in a breakdown of spermatocyte cyst structure and germ cell individualization. Specific dlg expression in SCCs resulted in developmental rescue of dlg mutant testes, whereas its expression in germ cells exerted no such effect. dlg overexpression in wt testes led to spermatocyte cyst expansion at the expense of spermatogonial cysts. Our data demonstrate that dlg is essentially required in SCCs for their survival, expansion, and differentiation, and for the encapsulation of the germline cells. PMID: [19546890] Back to Top |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figures for illustrating the function of this protein/gene |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function |
During embryonic development, some isoforms areessential for proper neuronal differentiation and organization.Required for cell polarity; maintenance of apicobasal polarity.Plays a critical role at septate junctions in cellular growthcontrol during larval development. The presence of a guanylatekinase domain suggests involvement in cellular adhesion as well assignal transduction to control cellular proliferation. Back to Top |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subcellular Location |
Cytoplasm. Cell membrane; Peripheralmembrane protein; Cytoplasmic side. Cytoplasm, cytoskeleton. Celljunction, septate junction. Note=Cytoskeleton- and membrane-associated. Located at the cytoplasmic face of the membrane in thecellular blastoderm and becomes associated with septate junctionswhich begin to form between epithelial cells at the time of dorsalclosure. In adult flies, located at the apical-lateral membraneboundary of epithelial cells. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tissue Specificity |
During the cellular blastoderm stage, isoformB, isoform F, isoform H, isoform I and isoform L expression islocalized to the cell borders. From stage 11 onwards, expressionis found predominantly in the developing nervous system: axonbundles in the ventral cord and the brain. Stage 14 and 15 embryosexhibit expression in the developing body wall muscle. Expressionin neuropil regions of the CNS and at NMJs persists through tolarval development. Other isoforms show expression in embryonicepithelial cells. In larvae, expression is seen as a belt aroundsalivary glands, imaginal disks and proventriculus. Expressed inadult reproductive tissues. In epithelia, coexpressed with scribthroughout development. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Ontology |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Interpro |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Pfam |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
SMART |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PROSITE |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PRINTS |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Created Date |
18-Oct-2012 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Record Type |
Experiment identified |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein sequence Annotation |
CHAIN 1 970 Disks large 1 tumor suppressor protein. /FTId=PRO_0000094538. DOMAIN 4 64 L27. DOMAIN 216 303 PDZ 1. DOMAIN 330 421 PDZ 2. DOMAIN 506 587 PDZ 3. DOMAIN 620 690 SH3. DOMAIN 780 955 Guanylate kinase-like. MOD_RES 496 496 Phosphoserine. MOD_RES 714 714 Phosphothreonine. VAR_SEQ 1 92 MPVKKQEAHRALELLEDYHARLSEPQDRALRIAIERVIRIF KSRLFQALLDIQEFYELTLLDDSKSIQQKTAETLQIATKWE KDGQAVKIAD -> MIDWVSIVRHSRRRFSNYVGSRSPVRM RRRRRQLTAPPPQQQQQQHYHQQQQQDQHQSRERQKKDKEK EKETEKDNESGGGIGSRYACCCAN (in isoform K). /FTId=VSP_039402. VAR_SEQ 1 37 MPVKKQEAHRALELLEDYHARLSEPQDRALRIAIERV -> MTTRKKKRDGGGSGGGFIKKVSSLFNLDSLHKASSTK (in isoform A). /FTId=VSP_011403. VAR_SEQ 1 29 MPVKKQEAHRALELLEDYHARLSEPQDRA -> MTTRKKKR DGGGSGGGFIKKVSSLFNLDS (in isoform E and isoform G). /FTId=VSP_011402. VAR_SEQ 1 7 MPVKKQE -> MDSDTDSEREKSSDPNEGLLSSDDKTFHDD DEPAEDSSPADDEEEPEEEECLLPQKKAQIRCDQDQPPLVV LVQPSAEAIEVRQEIDDTNPVAVAAKASDMDGDSQLEVMEH QMETVTEPDPEPPKCPTSLRDSVRESVECFYSAQDLLEYGH MLSSTSMVRTPDVESGYFEKSESDASRDEWEGPSSSSSGAA RCRLLSGISGLSVSSSSRHSAEGLRMELSRFRTMIETLERE SLEKSQSELQLKAKSKAKPKPKQRSHVQDAAGESGSEQGSE RGFWSTIFGQAGLAISQDEEERIADIQK (in isoform F). /FTId=VSP_011401. VAR_SEQ 30 205 Missing (in isoform E and isoform G). /FTId=VSP_011404. VAR_SEQ 38 205 Missing (in isoform A). /FTId=VSP_011405. VAR_SEQ 93 151 Missing (in isoform I, isoform H, isoform K and isoform L). /FTId=VSP_011406. VAR_SEQ 205 205 T -> TLHKASSTK (in isoform L). /FTId=VSP_011407. VAR_SEQ 206 267 VNGDDSWLYEDIQLERGNSGLGFSIAGGTDNPHIGTDTSIY ITKLISGGAAAADGRLSINDI -> SQIQIQSLTQTYPNAH QRKRVLVSLHPHQHQHQSQIQHQHHYQLRHNNGIQAKMLKR AFEST (in isoform I). /FTId=VSP_011408. VAR_SEQ 268 970 Missing (in isoform I). /FTId=VSP_011409. VAR_SEQ 473 519 EPGSRYASTNVLAAVPPGTPRAVSTEDITREPRTITIQKGP QGLGFN -> AFMLCYTQDDANAEGGEIIYRVELPDMEQIT LIYLENNDADYRKSSI (in isoform F). /FTId=VSP_011411. VAR_SEQ 473 473 E -> GALNSMGQTVVDSPSIPQAAAAVAAAANASASASVI ASNNTISNTTVTTVTATATASNSSSKLPPSLGANSSISISN SNSNSNSNNINNINSINNNNSSSSSTTATVAAATPTAASAA AAAASSPPANSFYNNASMPALPVESNQTNNRSQSPQPRQ (in isoform A, isoform E and isoform G). /FTId=VSP_011410. VAR_SEQ 520 970 Missing (in isoform F). /FTId=VSP_011412. VAR_SEQ 746 746 P -> PNGVVSSTSEIDINNVNNNQSNEPQP (in isoform G). /FTId=VSP_011413. VAR_SEQ 747 761 FMLCYTQDDANAEGA -> NGVVSSTSEIDINNVNNNQSNE PQP (in isoform A and isoform E). /FTId=VSP_011414. VAR_SEQ 761 761 A -> GEIIYRVELPDMEQITLIYLENNDADYP (in isoform L). /FTId=VSP_011415. CONFLICT 365 365 M -> T (in Ref. 1; AAA28468 and 2; AAQ01226). CONFLICT 369 369 A -> R (in Ref. 1; AAA28468 and 2; AAQ01226). CONFLICT 395 395 E -> G (in Ref. 6; ACV53090). Back to Top |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Nucleotide Sequence |
Length: 3263 bp Go to nucleotide: FASTA |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Sequence |
Length: 970 bp Go to amino acid: FASTA |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The verified Protein-Protein interaction information |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Comments |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||