| Tag | Content | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00000151 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
UniProt Accession |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Theoretical PI |
11.35
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Weight |
31031 Da
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genbank Nucleotide ID |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genbank Protein ID |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Name |
tra2 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Synonyms/Alias |
tra-2 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Name |
Transformer-2 sex-determining protein |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Synonyms/Alias |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Organism |
Drosophila melanogaster (Fruit fly) |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
NCBI Taxonomy ID |
7227 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Chromosome Location |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function in Stage |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function in Cell Type |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Description |
Temporarily unavailable |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The information of related literatures |
1. M. E. McGuffin, D. Chandler, D. Somaiya, B. Dauwalder and W. Mattox (1998) Autoregulation of transformer-2 alternative splicing is necessary for normal male fertility in Drosophila. Genetics 149(3): 1477-86. Abstract In the male germline of Drosophila the transformer-2 protein is required for differential splicing of pre-mRNAs from the exuperantia and att genes and autoregulates alternative splicing of its own pre-mRNA. Autoregulation of TRA-2 splicing results in production of two mRNAs that differ by the splicing/retention of the M1 intron and encode functionally distinct protein isoforms. Splicing of the intron produces an mRNA encoding TRA-2(226), which is necessary and sufficient for both male fertility and regulation of downstream target RNAs. When the intron is retained, an mRNA is produced encoding TRA-2(179), a protein with no known function. We have previously shown that repression of M1 splicing is dependent on TRA-2(226), suggesting that this protein quantitatively limits its own expression through a negative feedback mechanism at the level of splicing. Here we examine this idea, by testing the effect that variations in the level of tra-2 expression have on the splicing of M1 and on male fertility. Consistent with our hypothesis, we observe that as tra-2 gene dosage is increased, smaller proportions of TRA-2(226) mRNA are produced, limiting expression of this isoform. Feedback regulation is critical for male fertility, since it is significantly decreased by a transgene in which repression of M1 splicing cannot occur and TRA-2(226) mRNA is constitutively produced. The effect of this transgene becomes more severe as its dosage is increased, indicating that fertility is sensitive to an excess of TRA-2(226). Our results suggest that autoregulation of TRA-2(226) expression in male germ cells is necessary for normal spermatogenesis. PMID: [9649535] 2. W. Mattox, M. E. McGuffin and B. S. Baker (1996) A negative feedback mechanism revealed by functional analysis of the alternative isoforms of the Drosophila splicing regulator transformer-2. Genetics 143(1): 303-14. Abstract The Drosophila sex determination gene transformer-2 (tra-2) is a splicing regulator that affects the sex-specific processing of several distinct pre-mRNAs. While the tra-2 gene itself is known to produce alternative mRNAs that together encode three different TRA-2 protein isoforms, the respective roles of these isoforms in affecting individual pre-mRNA targets has remained unclear. We have generated transgenic fly strains with mutations affecting specific TRA-2 isoforms to investigate their individual roles in regulating the alternative processing of doublesex, exuperantia and tra-2 pre-mRNA. Our results indicate that in somatic tissues two different isoforms function redundantly to direct female differentiation and female-specific doublesex pre-mRNA splicing. In the male germline, where tra-2 has an essential role in spermatogenesis, a single isoform was found to uniquely perform all necessary functions. This isoform appears to regulate its own synthesis during spermatogenesis through a negative feedback mechanism involving intron retention. PMID: [8722783] Back to Top |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
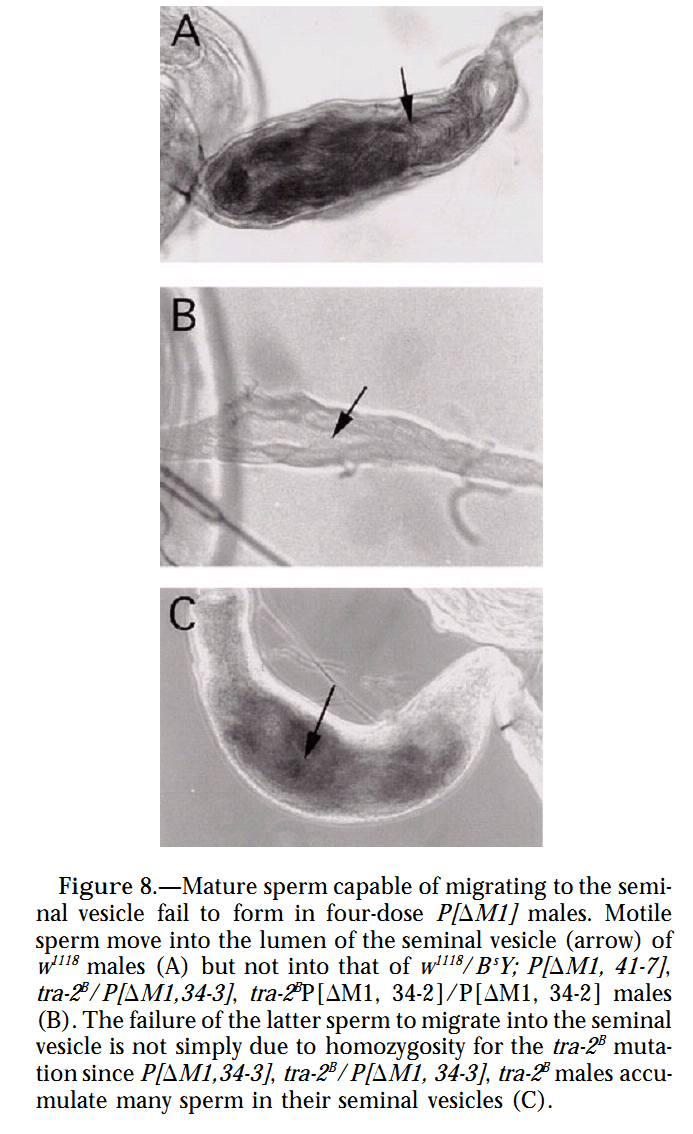
Figures for illustrating the function of this protein/gene |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function |
Required for female sex determination in somatic cellsand for spermatogenesis in male germ cells. Positive regulator offemale-specific splicing and/or polyadenylation of doublesex (dsx)pre-mRNA. Splicing requires an enhancer complex, dsxRE (dsx repeatelement: which contains six copies of a 13-nucleotide repeat and apurine-rich enhancer (PRE)). DsxRE is formed through cooperativeinteractions between tra, tra2 and the sr proteins, and theseinteractions require both the repeat sequences and PRE. PRE isrequired for specific binding of tra2 to the dsxRE. Protein-RNAand protein-protein interactions are involved in tra-2 dependentactivation and repression of alternative splicing. Together withtra-2, plays a role in switching fru splicing from the male-specific pattern to the female-specific pattern through activationof the female-specific fru 5'-splice site. Back to Top |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subcellular Location |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tissue Specificity |
Isoform Tmaj and isoform Tmin are expressed inmales and females. Isoform msTmaj and isoform msTmin are presentonly in male germ cells. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Ontology |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Interpro |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Pfam |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
SMART |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PROSITE |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PRINTS |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Created Date |
18-Oct-2012 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Record Type |
Experiment identified |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein sequence Annotation |
CHAIN 1 264 Transformer-2 sex-determining protein. /FTId=PRO_0000081986. DOMAIN 97 175 RRM. REGION 176 196 Linker. COMPBIAS 21 90 Arg/Ser-rich (RS1 domain). COMPBIAS 197 264 Arg/Ser-rich (RS2 domain). MOD_RES 40 40 Phosphoserine. MOD_RES 180 180 Phosphothreonine. MOD_RES 212 212 Phosphoserine. MOD_RES 214 214 Phosphoserine. MOD_RES 254 254 Phosphoserine. VAR_SEQ 1 85 Missing (in isoform MsTmaj). /FTId=VSP_005900. VAR_SEQ 2 39 Missing (in isoform Tmin). /FTId=VSP_005901. VAR_SEQ 134 264 TQRSRGFCFIYFEKLSDARAAKDSCSGIEVDGRRIRVDFSI TQRAHTPTPGVYLGRQPRGKAPRSFSPRRGRRVYHDRSASP YDNYRDRYDYRNDRYDRNLRRSPSRNRYTRNRSYSRSRSPQ LRRTSSRY -> INP (in isoform MsTmin). /FTId=VSP_005902. MUTAGEN 11 82 Missing: In d2; greatly reduced female- specific DSX splicing. MUTAGEN 49 82 Missing: In d1; greatly reduced female- specific DSX splicing. Retains male fertility. MUTAGEN 113 123 Missing: In a36; loss of female-specific DSX splicing. Loss of male fertility. MUTAGEN 138 138 R->L: In pm1; loss of female-specific DSX splicing. Loss of male fertility. MUTAGEN 140 140 F->A: In pm2; no female-specific DSX splicing. Some low male fertility. MUTAGEN 151 151 A->V: In ts1; little female-specific DSX splicing. Loss of male fertility and temperature-sensitive phenotype. MUTAGEN 172 264 Missing: In d5; loss of female-specific DSX splicing. Loss of male fertility. MUTAGEN 173 173 S->A: In pm3; greatly reduced female- specific DSX splicing. Retains male fertility and temperature-sensitive phenotype. MUTAGEN 173 173 S->F: In a15, loss of female-specific DSX splicing and male fertility. MUTAGEN 173 173 S->T: In pm4; loss of female-specific DSX splicing. Retains male fertility and temperature-sensitive phenotype. MUTAGEN 181 181 P->S: In ts2; temperature-sensitive phenotype. MUTAGEN 205 264 Missing: In d4; loss of female-specific DSX splicing. Greatly reduced male fertility. MUTAGEN 236 264 Missing: In d3; greatly reduced female- specific DSX splicing. Retains male fertility. Back to Top |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Nucleotide Sequence |
Length: 1261 bp Go to nucleotide: FASTA |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Sequence |
Length: 264 bp Go to amino acid: FASTA |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The verified Protein-Protein interaction information |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Comments |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||