| Tag | Content | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00000230 |
|||||||||||||||||||||||||||||||||||||||
UniProt Accession |
||||||||||||||||||||||||||||||||||||||||
Theoretical PI |
6.58
|
|||||||||||||||||||||||||||||||||||||||
Molecular Weight |
184624 Da
|
|||||||||||||||||||||||||||||||||||||||
Genbank Nucleotide ID |
||||||||||||||||||||||||||||||||||||||||
Genbank Protein ID |
||||||||||||||||||||||||||||||||||||||||
Gene Name |
pcm |
|||||||||||||||||||||||||||||||||||||||
Gene Synonyms/Alias |
ORFNames=Dmel_CG3291 |
|||||||||||||||||||||||||||||||||||||||
Protein Name |
||||||||||||||||||||||||||||||||||||||||
Protein Synonyms/Alias |
SubName: Pacman, isoform BEC=3.1.13.- |
|||||||||||||||||||||||||||||||||||||||
Organism |
Drosophila melanogaster (Fruit fly) |
|||||||||||||||||||||||||||||||||||||||
NCBI Taxonomy ID |
7227 |
|||||||||||||||||||||||||||||||||||||||
Chromosome Location |
|
|||||||||||||||||||||||||||||||||||||||
Function in Stage |
||||||||||||||||||||||||||||||||||||||||
Function in Cell Type |
||||||||||||||||||||||||||||||||||||||||
Description |
Temporarily unavailable |
|||||||||||||||||||||||||||||||||||||||
The information of related literatures |
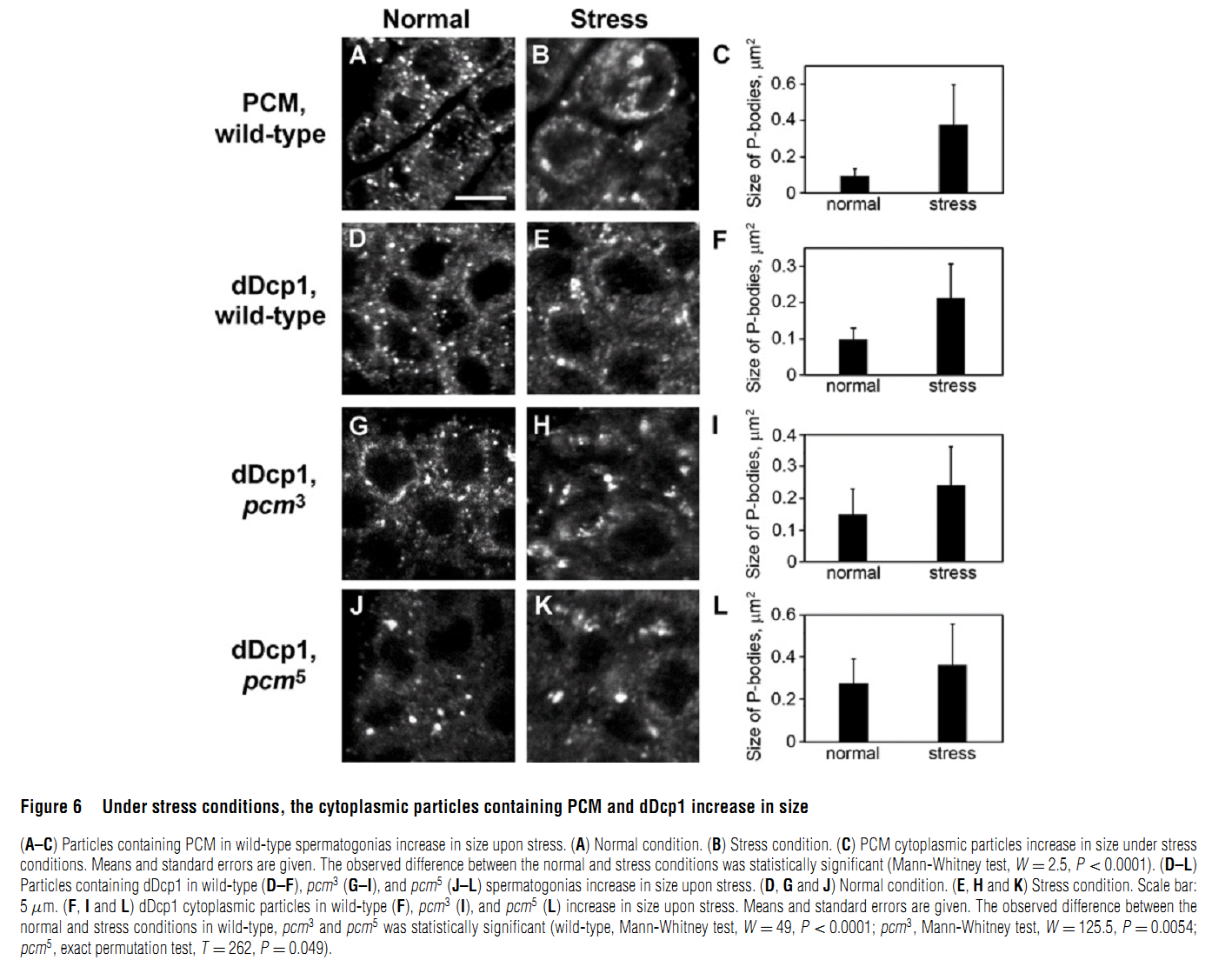
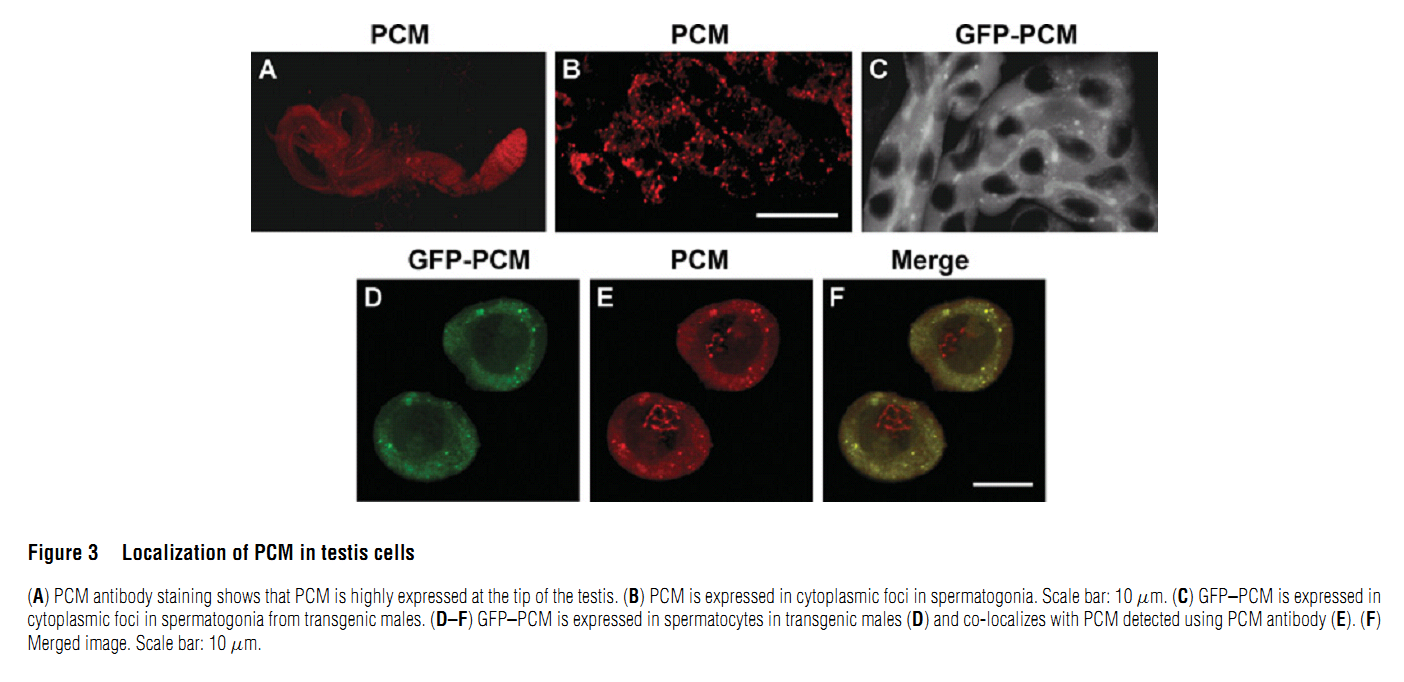
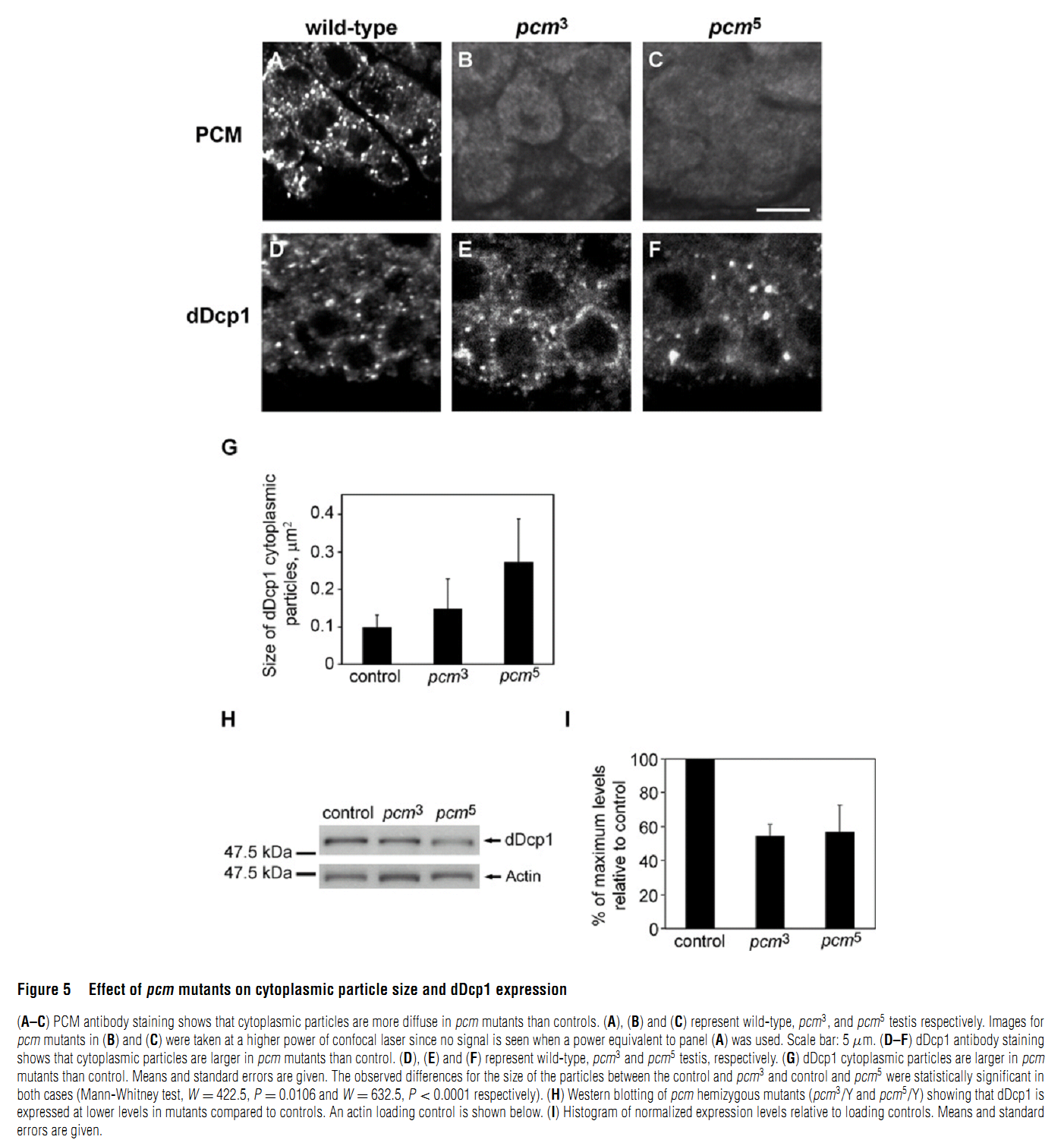
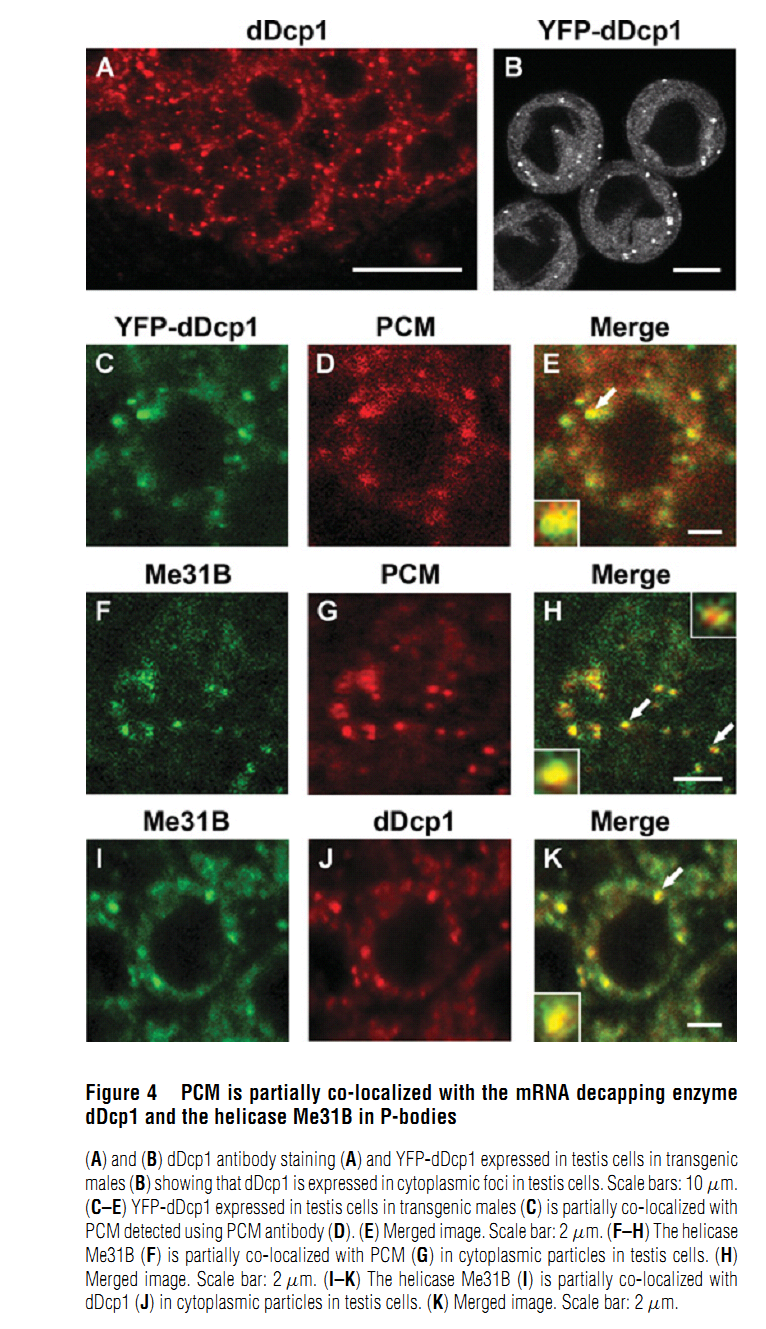
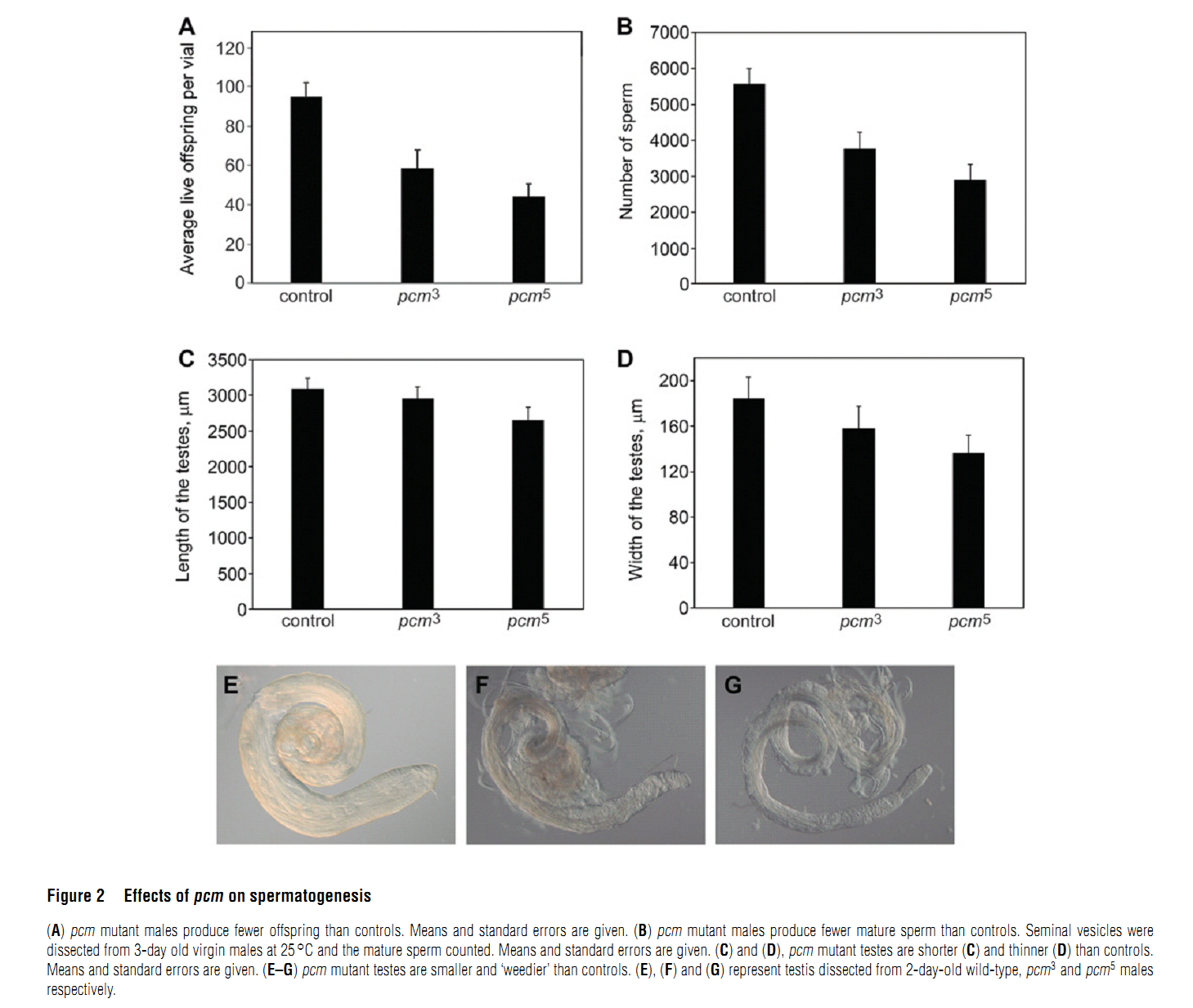
Abstract PMID: [18652575] 2. M. V. Zabolotskaya, D. P. Grima, M. D. Lin, T. B. Chou and S. F. Newbury (2008) The 5'-3' exoribonuclease Pacman is required for normal male fertility and is dynamically localized in cytoplasmic particles in Drosophila testis cells. Biochem J 416(3): 327-35. Abstract The exoribonuclease Xrn1 is widely recognised as a key component in the 5'-3' RNA degradation pathway. This enzyme is highly conserved between yeast and humans and is known to be involved in RNA interference and degradation of microRNAs as well as RNA turnover. In yeast and human tissue culture cells, Xrn1 has been shown to be a component of P-bodies (processing bodies), dynamic cytoplasmic granules where RNA degradation can take place. In this paper we show for the first time that Pacman, the Drosophila homologue of Xrn1, is localized in cytoplasmic particles in Drosophila testis cells. These particles are present in both the mitotically dividing spermatogonia derived from primordial stem cells and in the transcriptionally active spermatocytes. Pacman is co-localized with the decapping activator dDcp1 and the helicase Me31B (a Dhh1 homologue) in these particles, although this co-localization is not completely overlapping, suggesting that there are different compartments within these granules. Particles containing Pacman respond to stress and depletion of 5'-3' decay factors in the same way as yeast P-bodies, and therefore are likely to be sites of mRNA degradation or storage. Pacman is shown to be required for normal Drosophila spermatogenesis, suggesting that control of mRNA stability is crucial in the testis differentiation pathway. PMID: [18652574] Back to Top |
|||||||||||||||||||||||||||||||||||||||
Figures for illustrating the function of this protein/gene |
|
|||||||||||||||||||||||||||||||||||||||
Function |
||||||||||||||||||||||||||||||||||||||||
Subcellular Location |
||||||||||||||||||||||||||||||||||||||||
Tissue Specificity |
||||||||||||||||||||||||||||||||||||||||
Gene Ontology |
|
|||||||||||||||||||||||||||||||||||||||
Interpro |
||||||||||||||||||||||||||||||||||||||||
Pfam |
||||||||||||||||||||||||||||||||||||||||
SMART |
||||||||||||||||||||||||||||||||||||||||
PROSITE |
||||||||||||||||||||||||||||||||||||||||
PRINTS |
||||||||||||||||||||||||||||||||||||||||
Created Date |
18-Oct-2012 |
|||||||||||||||||||||||||||||||||||||||
Record Type |
Experiment identified |
|||||||||||||||||||||||||||||||||||||||
Protein sequence Annotation |
||||||||||||||||||||||||||||||||||||||||
Nucleotide Sequence |
Length: bp Go to nucleotide: FASTA |
|||||||||||||||||||||||||||||||||||||||
Protein Sequence |
Length: 1613 bp Go to amino acid: FASTA |
|||||||||||||||||||||||||||||||||||||||
The verified Protein-Protein interaction information |
| |||||||||||||||||||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
|||||||||||||||||||||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
|||||||||||||||||||||||||||||||||||||||
Comments |
||||||||||||||||||||||||||||||||||||||||