| Tag | Content | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00000280 |
|||||||||||||||||||||
UniProt Accession |
||||||||||||||||||||||
Theoretical PI |
5.3
|
|||||||||||||||||||||
Molecular Weight |
30088 Da
|
|||||||||||||||||||||
Genbank Nucleotide ID |
||||||||||||||||||||||
Genbank Protein ID |
||||||||||||||||||||||
Gene Name |
GN ORFNames=CG8979 |
|||||||||||||||||||||
Gene Synonyms/Alias |
||||||||||||||||||||||
Protein Name |
Putative proteasome inhibitor |
|||||||||||||||||||||
Protein Synonyms/Alias |
||||||||||||||||||||||
Organism |
Drosophila melanogaster (Fruit fly) |
|||||||||||||||||||||
NCBI Taxonomy ID |
7227 |
|||||||||||||||||||||
Chromosome Location |
|
|||||||||||||||||||||
Function in Stage |
||||||||||||||||||||||
Function in Cell Type |
||||||||||||||||||||||
Description |
Temporarily unavailable |
|||||||||||||||||||||
The information of related literatures |
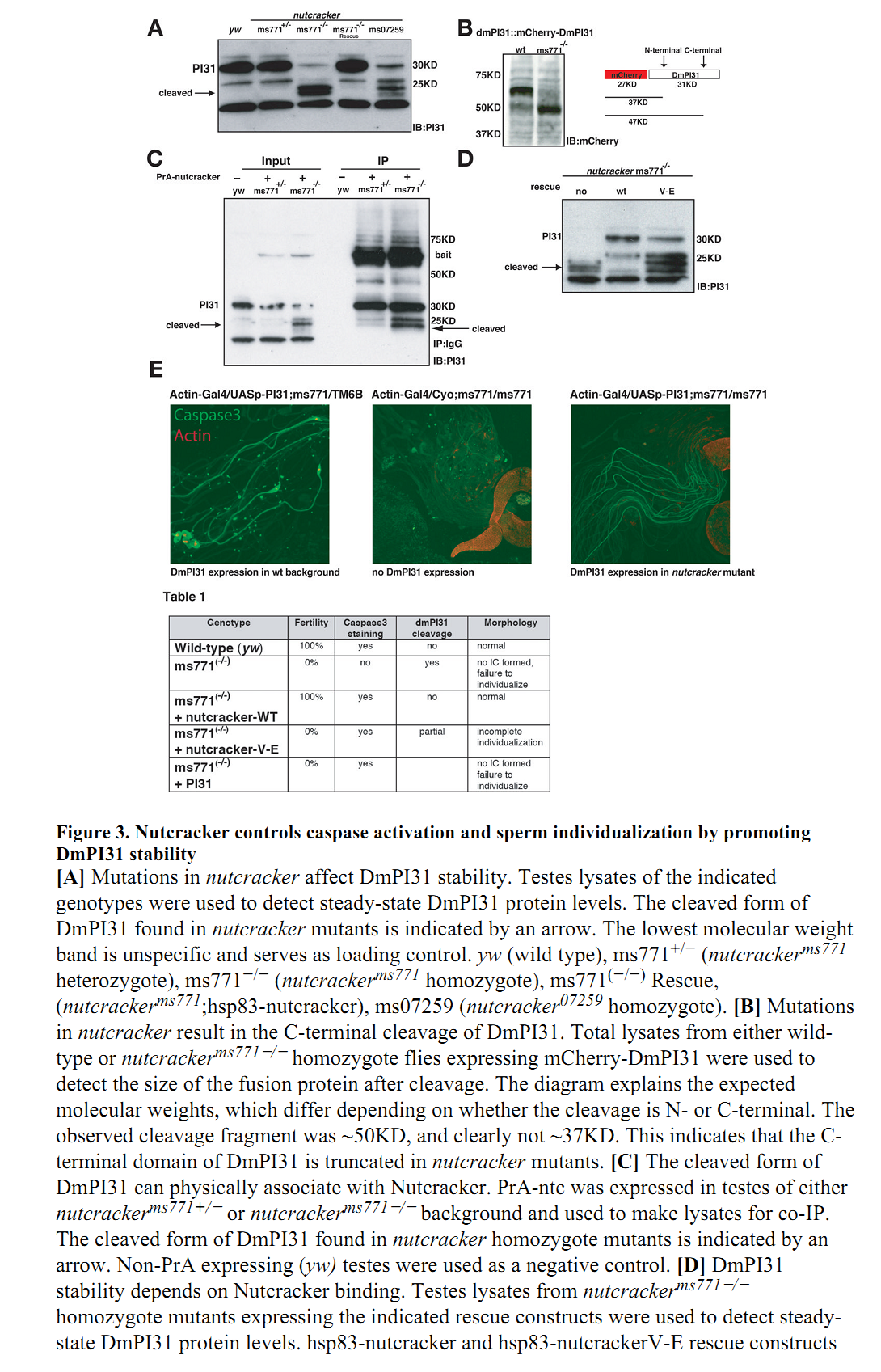
1. M. Bader, S. Benjamin, O. L. Wapinski, D. M. Smith, A. L. Goldberg and H. Steller (2011) A conserved F box regulatory complex controls proteasome activity in Drosophila. Cell 145(3): 371-82. Abstract The ubiquitin-proteasome system catalyzes the degradation of intracellular proteins. Although ubiquitination of proteins determines their stabilities, there is growing evidence that proteasome function is also regulated. We report the functional characterization of a conserved proteasomal regulatory complex. We identified DmPI31 as a binding partner of the F box protein Nutcracker, a component of an SCF ubiquitin ligase (E3) required for caspase activation during sperm differentiation in Drosophila. DmPI31 binds Nutcracker via a conserved mechanism that is also used by mammalian FBXO7 and PI31. Nutcracker promotes DmPI31 stability, which is necessary for caspase activation, proteasome function, and sperm differentiation. DmPI31 can activate 26S proteasomes in vitro, and increasing DmPI31 levels suppresses defects caused by diminished proteasome activity in vivo. Furthermore, loss of DmPI31 function causes lethality, cell-cycle abnormalities, and defects in protein degradation, demonstrating that DmPI31 is physiologically required for normal proteasome activity. PMID: [21529711] Back to Top |
|||||||||||||||||||||
Figures for illustrating the function of this protein/gene |
|
|||||||||||||||||||||
Function |
Could play an important role in control of proteasomefunction. Inhibits the hydrolysis of protein and peptidesubstrates by the 20S proteasome (By similarity). Back to Top |
|||||||||||||||||||||
Subcellular Location |
||||||||||||||||||||||
Tissue Specificity |
||||||||||||||||||||||
Gene Ontology |
|
|||||||||||||||||||||
Interpro |
||||||||||||||||||||||
Pfam |
||||||||||||||||||||||
SMART |
||||||||||||||||||||||
PROSITE |
||||||||||||||||||||||
PRINTS |
||||||||||||||||||||||
Created Date |
18-Oct-2012 |
|||||||||||||||||||||
Record Type |
Experiment identified |
|||||||||||||||||||||
Protein sequence Annotation |
CHAIN 1 270 Putative proteasome inhibitor. /FTId=PRO_0000220924. COMPBIAS 169 270 Pro-rich. MOD_RES 168 168 Phosphoserine. MOD_RES 189 189 Phosphoserine. Back to Top |
|||||||||||||||||||||
Nucleotide Sequence |
Length: bp Go to nucleotide: FASTA |
|||||||||||||||||||||
Protein Sequence |
Length: 270 bp Go to amino acid: FASTA |
|||||||||||||||||||||
The verified Protein-Protein interaction information |
||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
|||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
|||||||||||||||||||||
Comments |
||||||||||||||||||||||