| Tag | Content | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00000281 |
||||||||||||||||||||
UniProt Accession |
|||||||||||||||||||||
Theoretical PI |
5.06
|
||||||||||||||||||||
Molecular Weight |
58286 Da
|
||||||||||||||||||||
Genbank Nucleotide ID |
|||||||||||||||||||||
Genbank Protein ID |
|||||||||||||||||||||
Gene Name |
vis |
||||||||||||||||||||
Gene Synonyms/Alias |
ORFNames=Dmel_CG8821 |
||||||||||||||||||||
Protein Name |
|||||||||||||||||||||
Protein Synonyms/Alias |
SubName: Vismay, isoform B |
||||||||||||||||||||
Organism |
Drosophila melanogaster (Fruit fly) |
||||||||||||||||||||
NCBI Taxonomy ID |
7227 |
||||||||||||||||||||
Chromosome Location |
|
||||||||||||||||||||
Function in Stage |
|||||||||||||||||||||
Function in Cell Type |
|||||||||||||||||||||
Description |
Temporarily unavailable |
||||||||||||||||||||
The information of related literatures |
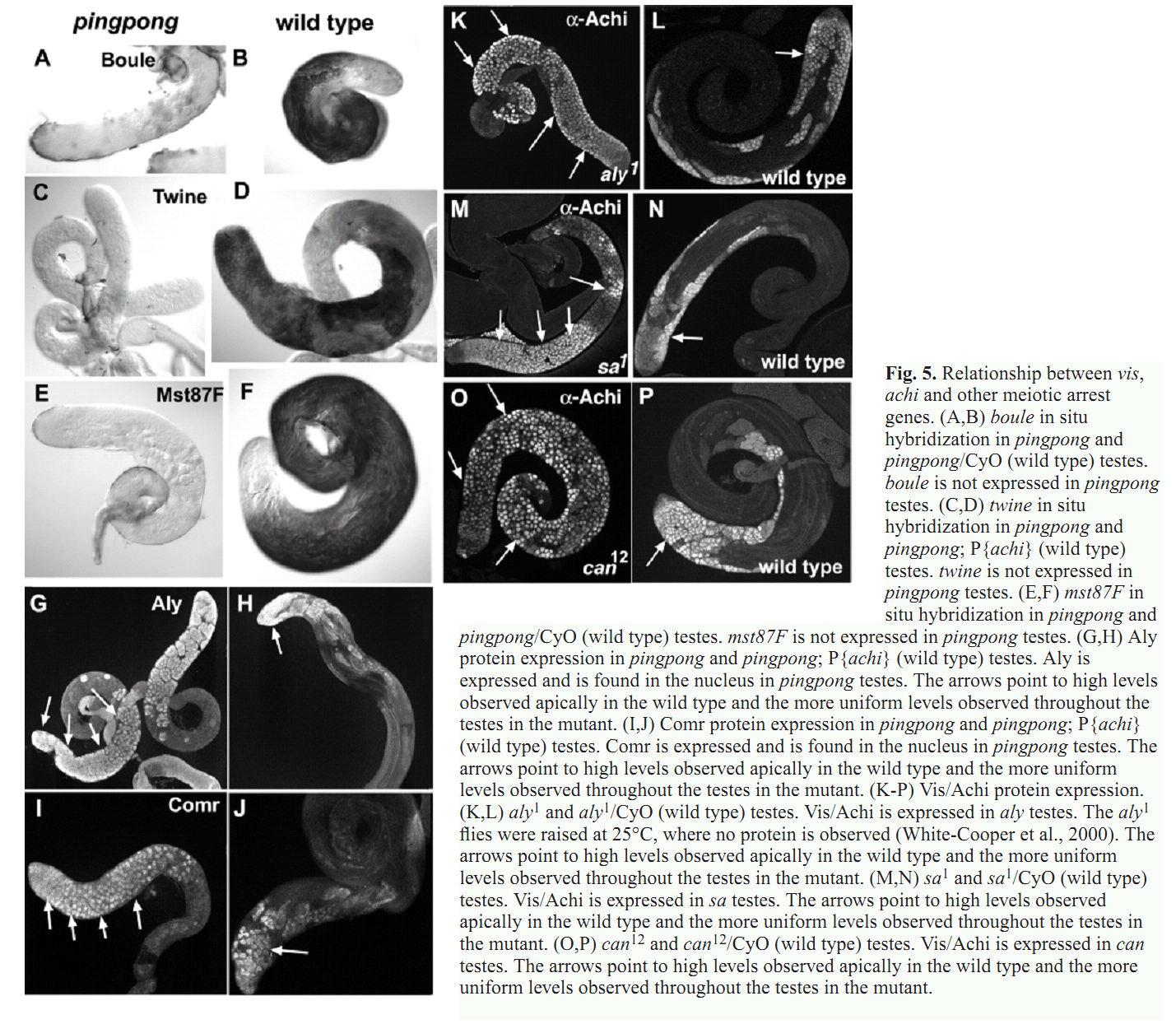
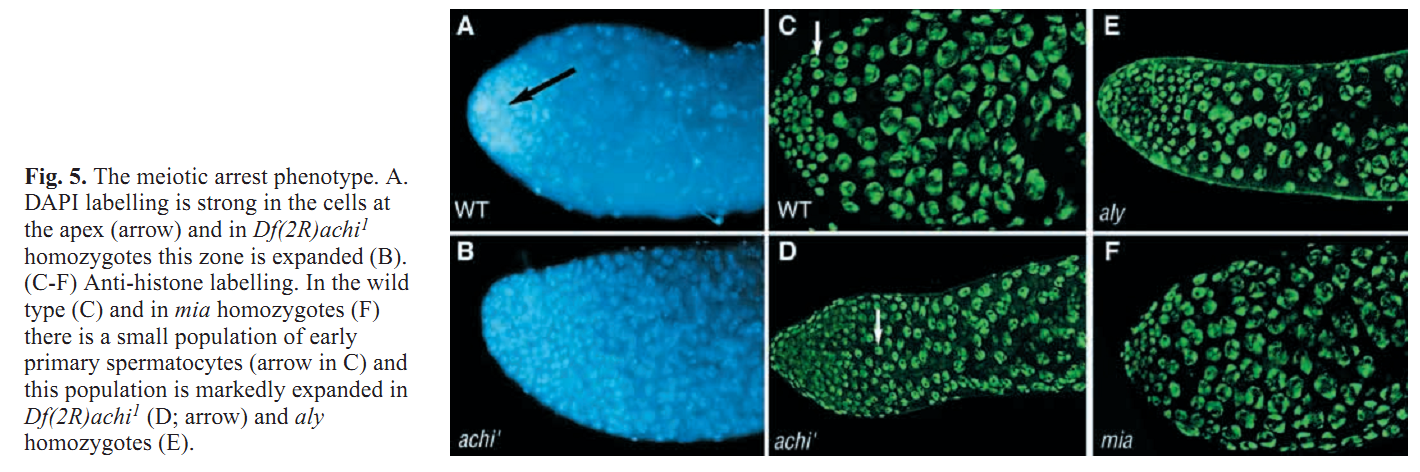
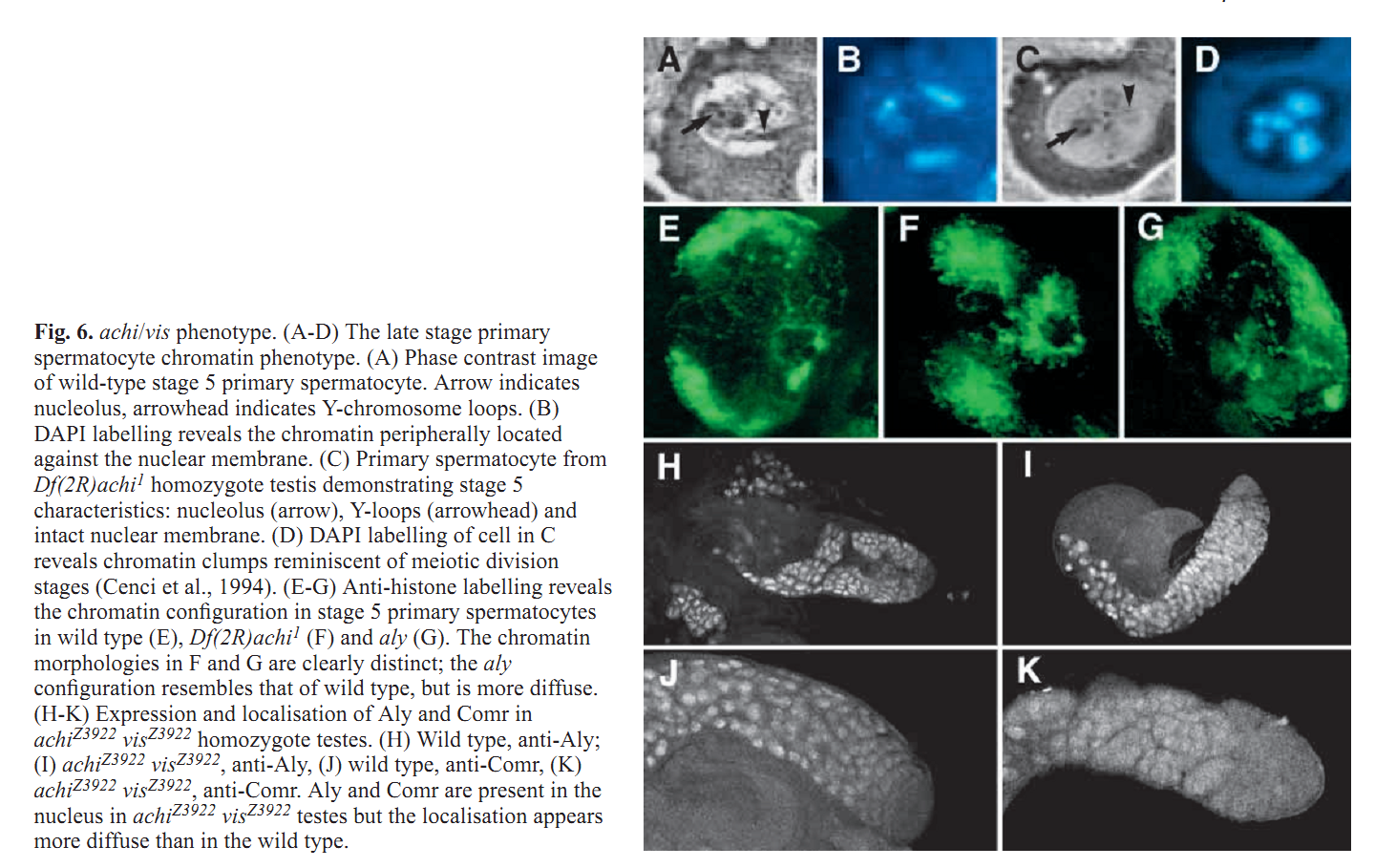
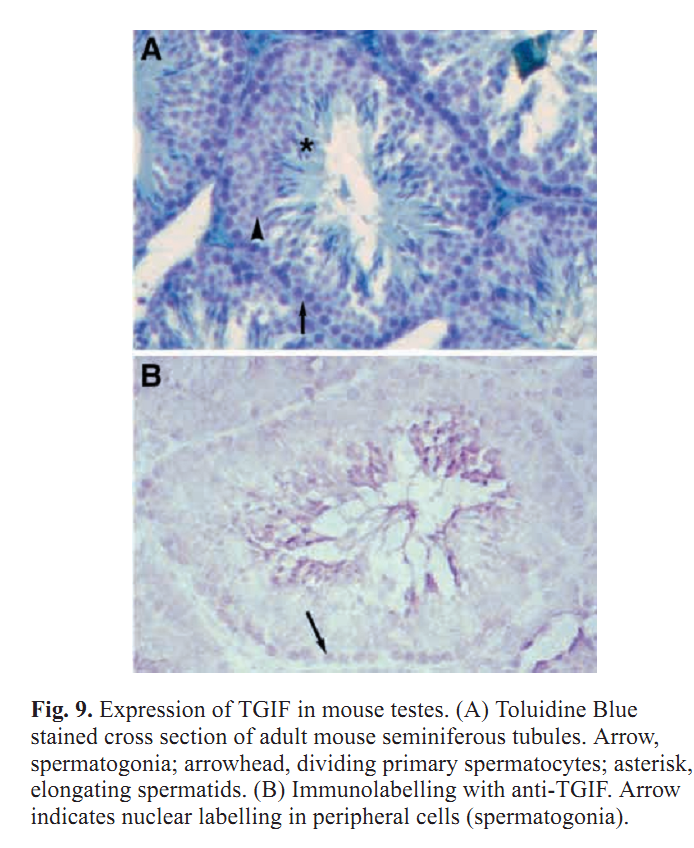
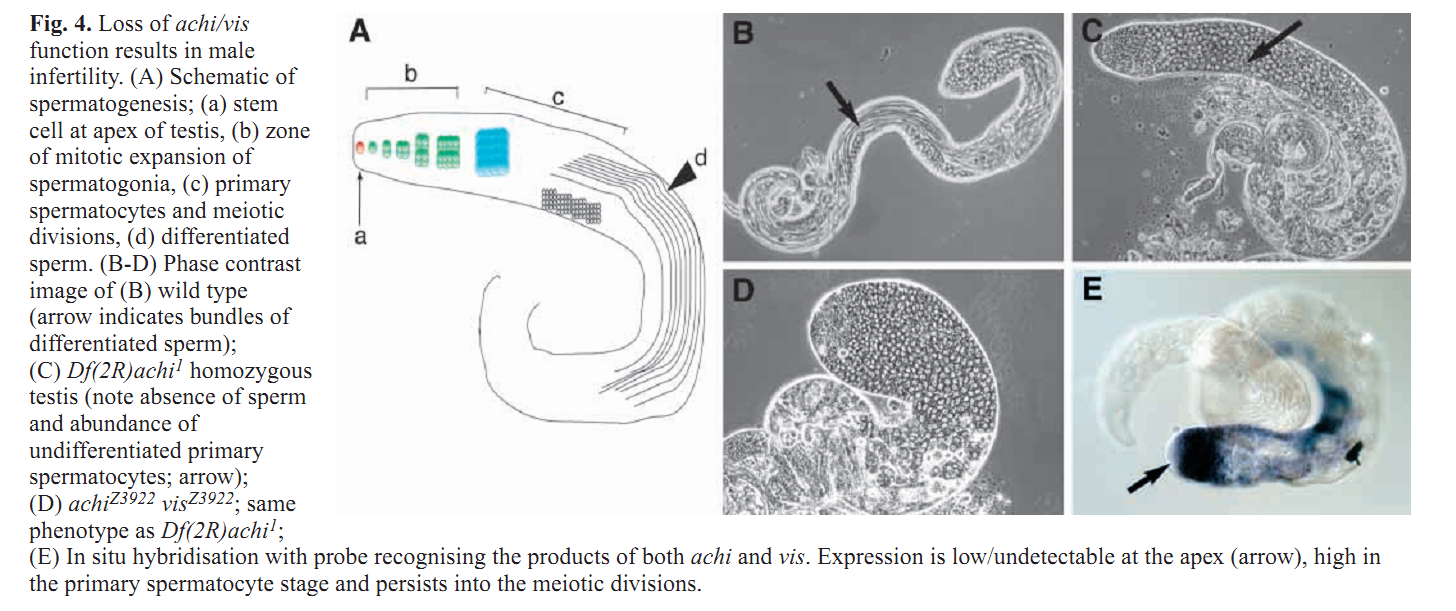
1. Z. Wang and R. S. Mann (2003) Requirement for two nearly identical TGIF-related homeobox genes in Drosophila spermatogenesis. Development 130(13): 2853-65. Abstract The genetic analysis of spermatogenesis in Drosophila melanogaster has led to the identification of several genes that control the onset of meiosis, spermatid differentiation, or both. We described two tightly linked and nearly identical homeobox genes of the TGIF (TG-interacting factor) subclass called vismay and achintya that are essential for spermatogenesis in Drosophila. In flies deficient for both genes, spermatogenesis is blocked prior to any spermatid differentiation and before the first meiotic division. This suggests that vismay and achintya function at the same step as two previously characterized meiotic arrest genes, always early and cookie monster. Consistent with this idea, both always early and cookie monster are still expressed in flies deficient in vismay and achintya. Conversely, Vismay and Achintya proteins are present in always early mutant testes. Co-immunoprecipitation experiments further suggest that Vismay and Achintya proteins exist in a complex with Always early and Cookie monster proteins. Because Vismay and Achintya are likely to be sequence-specific DNA binding factors, these results suggest that they help to specify the spermatogenesis program by recruiting or stabilizing Always early and Cookie monster to specific target genes that need to be transcriptionally regulated during testes development. PMID: [12756170] 2. J. C. Caldwell, M. L. Joiner, E. Sivan-Loukianova and D. F. Eberl (2008) The role of the RING-finger protein Elfless in Drosophila spermatogenesis and apoptosis. Fly (Austin) 2(6): 269-79. Abstract elfless (CG15150, FBgn0032660) maps to polytene region 36DE 5' (left) of reduced ocelli/Pray for Elves (PFE) on chromosome 2L and is predicted to encode a 187 amino acid RING finger E3 ubiquitin ligase that is putatively involved in programmed cell death (PCD, e.g., apoptosis). Several experimental approaches were used to characterize CG15150/elfless and test whether defects in this gene underlie the male sterile phenotype associated with overlapping chromosomal deficiencies of region 36DE. elfless expression is greatly enhanced in the testes and the expression pattern of UAS-elfless-EGFP driven by elfless-Gal4 is restricted to the tail cyst cell nuclei of the testes. Despite this, elfless transgenes failed to rescue the male sterile phenotype in Df/Df flies. Furthermore, null alleles of elfless, generated either by imprecise excision of an upstream P-element or by FLP-FRT deletion between two flanking piggyBac elements, are fertile. In a gain-of-function setting in the eye, we found that elfless genetically interacts with key members of the apoptotic pathway including the initiator caspase Dronc and the ubiquitin conjugating enzyme UbcD1. DIAP1, but not UbcD1, protein levels are increased in heads of flies expressing Elfless-EGFP in the eye, and in testes of flies expressing elfless-Gal4 driven Elfless-EGFP. Based on these findings, we speculate that Elfless may regulate tail cyst cell degradation to provide an advantageous, though not essential, function in the testis. PMID: [19077536] 3. S. Ayyar, J. Jiang, A. Collu, H. White-Cooper and R. A. White (2003) Drosophila TGIF is essential for developmentally regulated transcription in spermatogenesis. Development 130(13): 2841-52. Abstract We have investigated the role of TGIF, a TALE-class homeodomain transcription factor, in Drosophila development. In vertebrates, TGIF has been implicated, by in vitro analysis, in several pathways, most notably as a repressor modulating the response to TGFbeta signalling. Human TGIF has been associated with the developmental disorder holoprosencephaly. Drosophila TGIF is represented by the products of two tandemly repeated highly similar genes, achintya and vismay. We have generated mutations that delete both genes. Homozygous mutant flies are viable and appear morphologically normal, but the males are completely sterile. The defect lies at the primary spermatocyte stage and differentiation is blocked prior to the onset of the meiotic divisions. We show that mutants lacking TGIF function fail to activate transcription of many genes required for sperm manufacture and of some genes required for entry into the meiotic divisions. This groups TGIF together with two other genes producing similar phenotypes, always early and cookie monster, as components of the machinery required for the activation of the spermatogenic programme of transcription. TGIF is the first sequence-specific transcription factor identified in this pathway. By immunolabelling in mouse testes we show that TGIF is expressed in the early stages of spermatogenesis consistent with a conserved role in the activation of the spermatogenesis transcription programme. PMID: [12756169] Back to Top |
||||||||||||||||||||
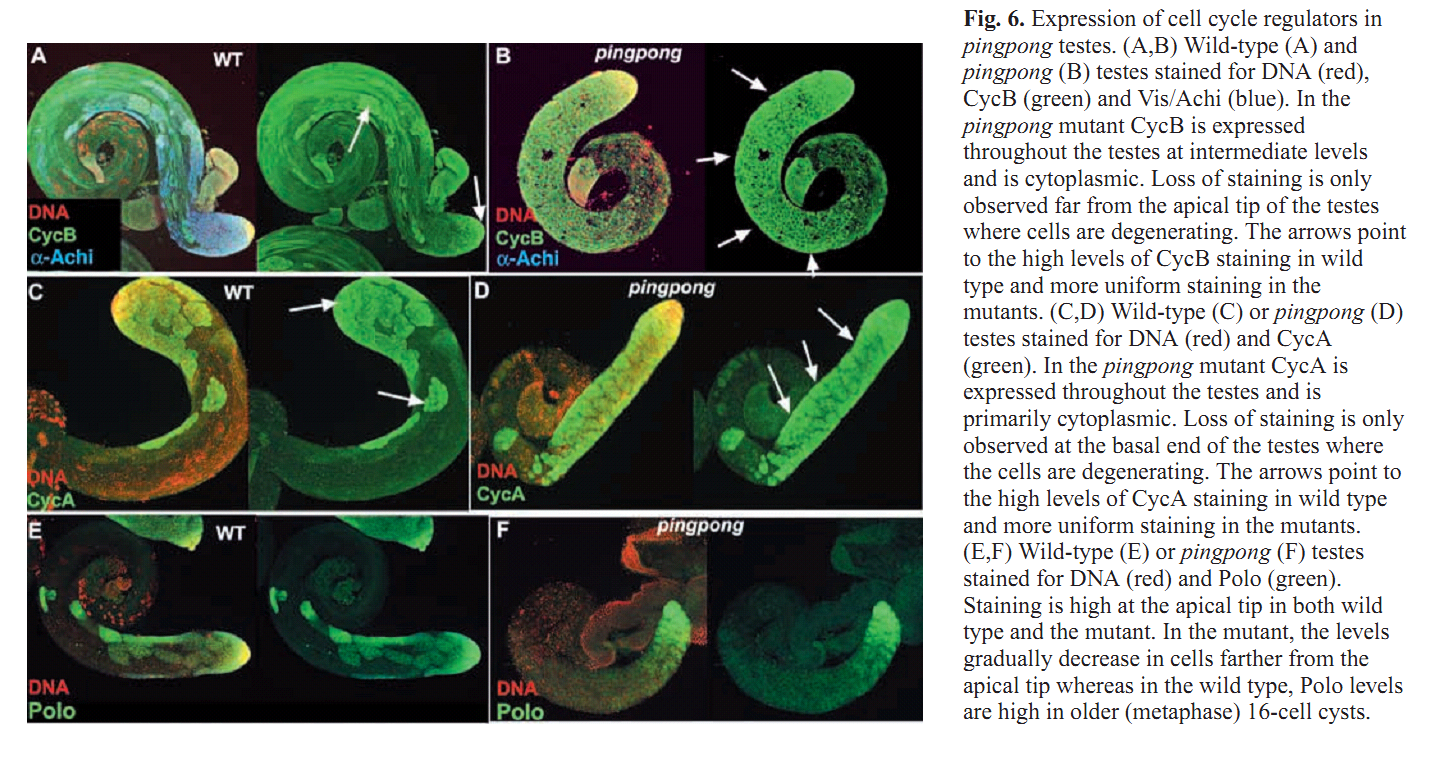
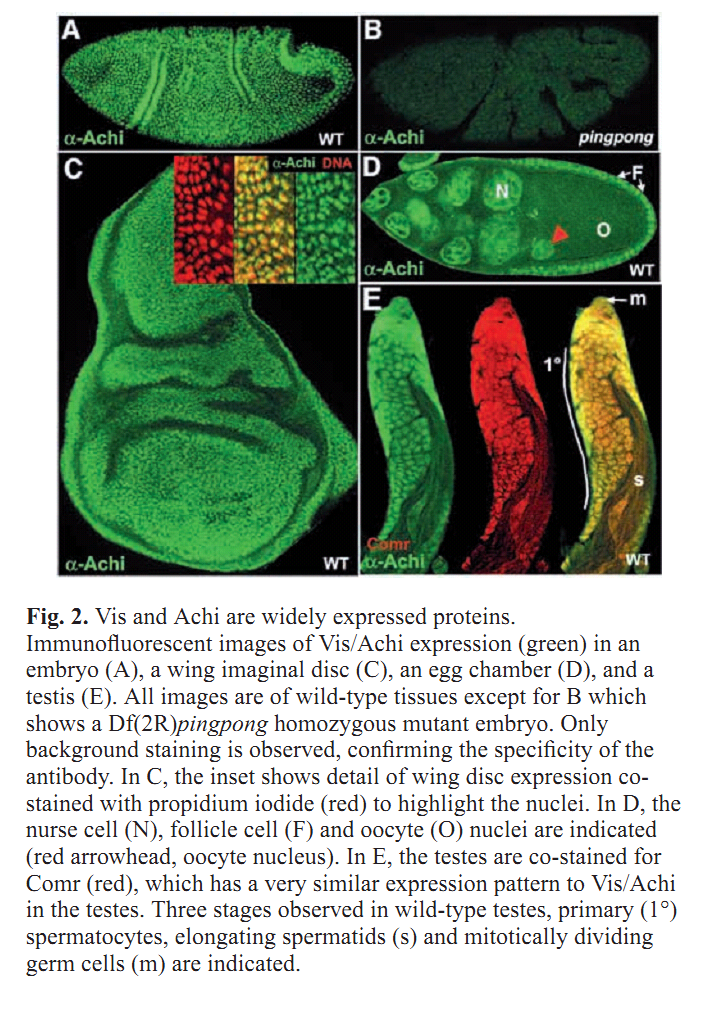
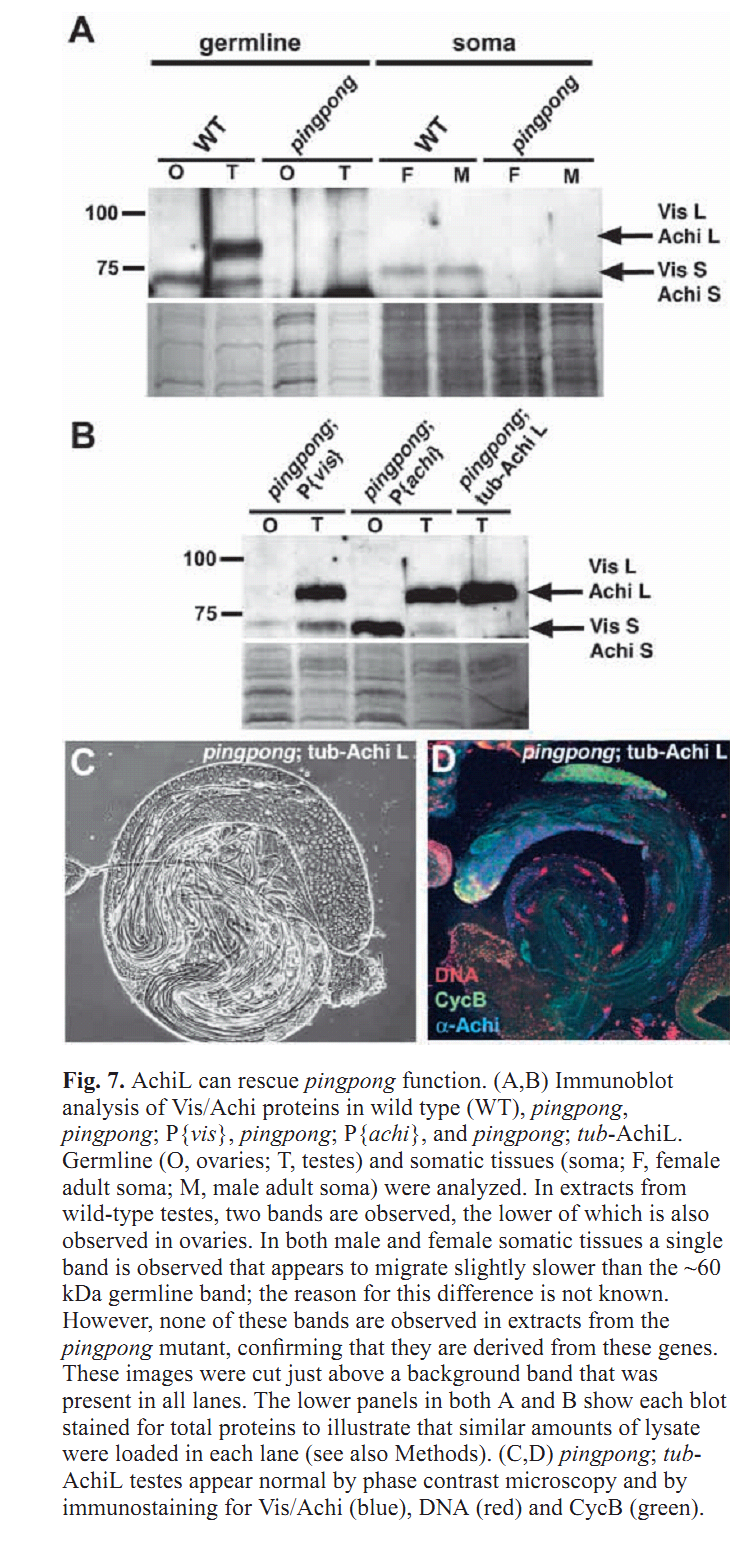
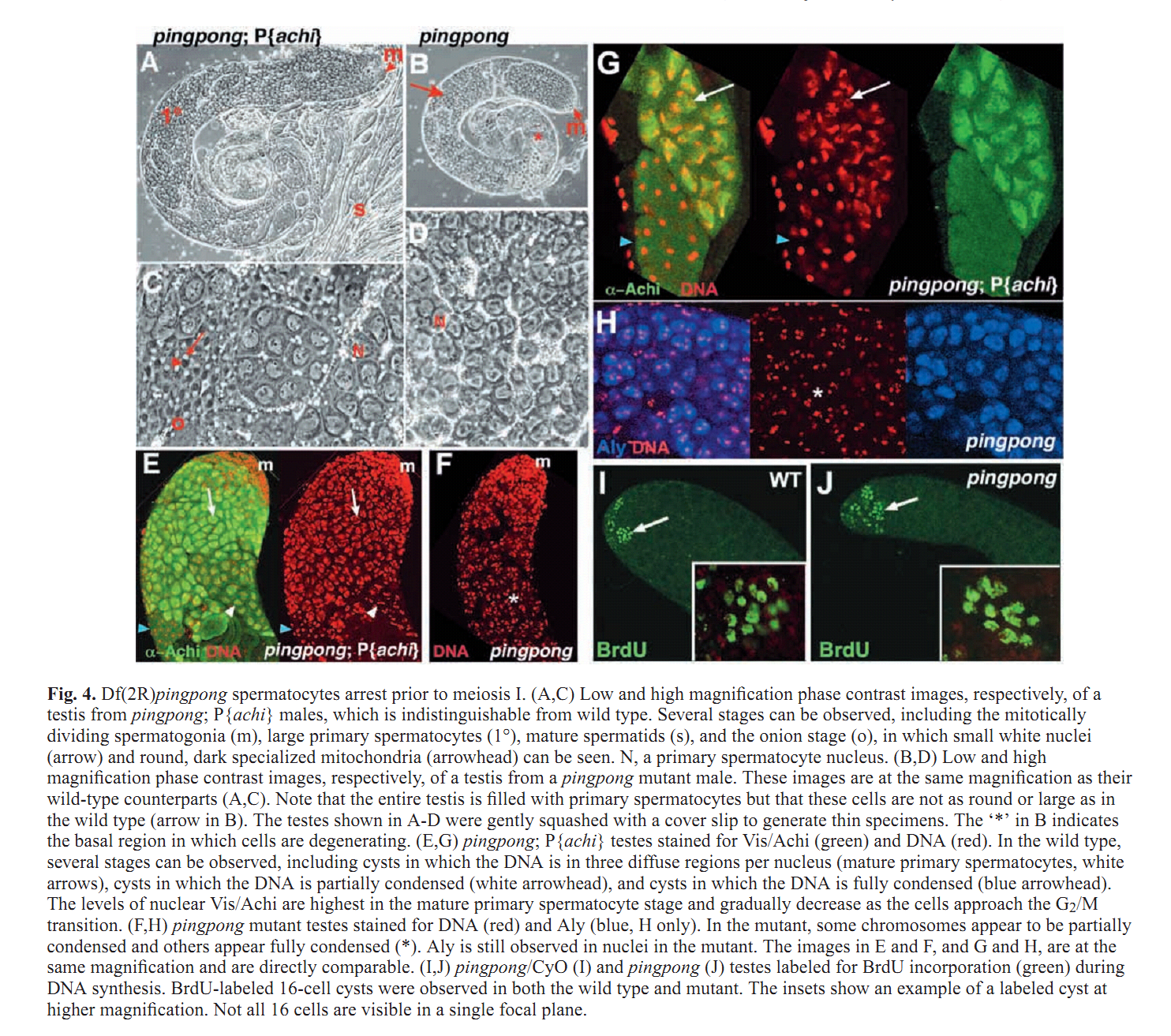
Figures for illustrating the function of this protein/gene |
|
||||||||||||||||||||
Function |
|||||||||||||||||||||
Subcellular Location |
Nucleus (By similarity).-----------------------------------------------------------------------Copyrighted by the UniProt Consortium, see http://www.uniprot.org/termsDistributed under the Creative Commons Attribution-NoDerivs License----------------------------------------------------------------------- |
||||||||||||||||||||
Tissue Specificity |
|||||||||||||||||||||
Gene Ontology |
|
||||||||||||||||||||
Interpro |
|||||||||||||||||||||
Pfam |
|||||||||||||||||||||
SMART |
|||||||||||||||||||||
PROSITE |
|||||||||||||||||||||
PRINTS |
|||||||||||||||||||||
Created Date |
18-Oct-2012 |
||||||||||||||||||||
Record Type |
Experiment identified |
||||||||||||||||||||
Protein sequence Annotation |
|||||||||||||||||||||
Nucleotide Sequence |
Length: bp Go to nucleotide: FASTA |
||||||||||||||||||||
Protein Sequence |
Length: 524 bp Go to amino acid: FASTA |
||||||||||||||||||||
The verified Protein-Protein interaction information |
| ||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
||||||||||||||||||||
Comments |
|||||||||||||||||||||