| Tag | Content | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00000313 |
||||||||||||||||||||||||||||||||||||||||||
UniProt Accession |
|||||||||||||||||||||||||||||||||||||||||||
Theoretical PI |
5.78
|
||||||||||||||||||||||||||||||||||||||||||
Molecular Weight |
54105 Da
|
||||||||||||||||||||||||||||||||||||||||||
Genbank Nucleotide ID |
|||||||||||||||||||||||||||||||||||||||||||
Genbank Protein ID |
|||||||||||||||||||||||||||||||||||||||||||
Gene Name |
park |
||||||||||||||||||||||||||||||||||||||||||
Gene Synonyms/Alias |
park-RC, parkin |
||||||||||||||||||||||||||||||||||||||||||
Protein Name |
|||||||||||||||||||||||||||||||||||||||||||
Protein Synonyms/Alias |
SubName: FI05213pSubName: PARKINSubName: Parkin, isoform BEC=6.3.2.19SubName: Parkin, isoform CEC=6.3.2.19 |
||||||||||||||||||||||||||||||||||||||||||
Organism |
Drosophila melanogaster (Fruit fly) |
||||||||||||||||||||||||||||||||||||||||||
NCBI Taxonomy ID |
7227 |
||||||||||||||||||||||||||||||||||||||||||
Chromosome Location |
|
||||||||||||||||||||||||||||||||||||||||||
Function in Stage |
|||||||||||||||||||||||||||||||||||||||||||
Function in Cell Type |
|||||||||||||||||||||||||||||||||||||||||||
Description |
Temporarily unavailable |
||||||||||||||||||||||||||||||||||||||||||
The information of related literatures |
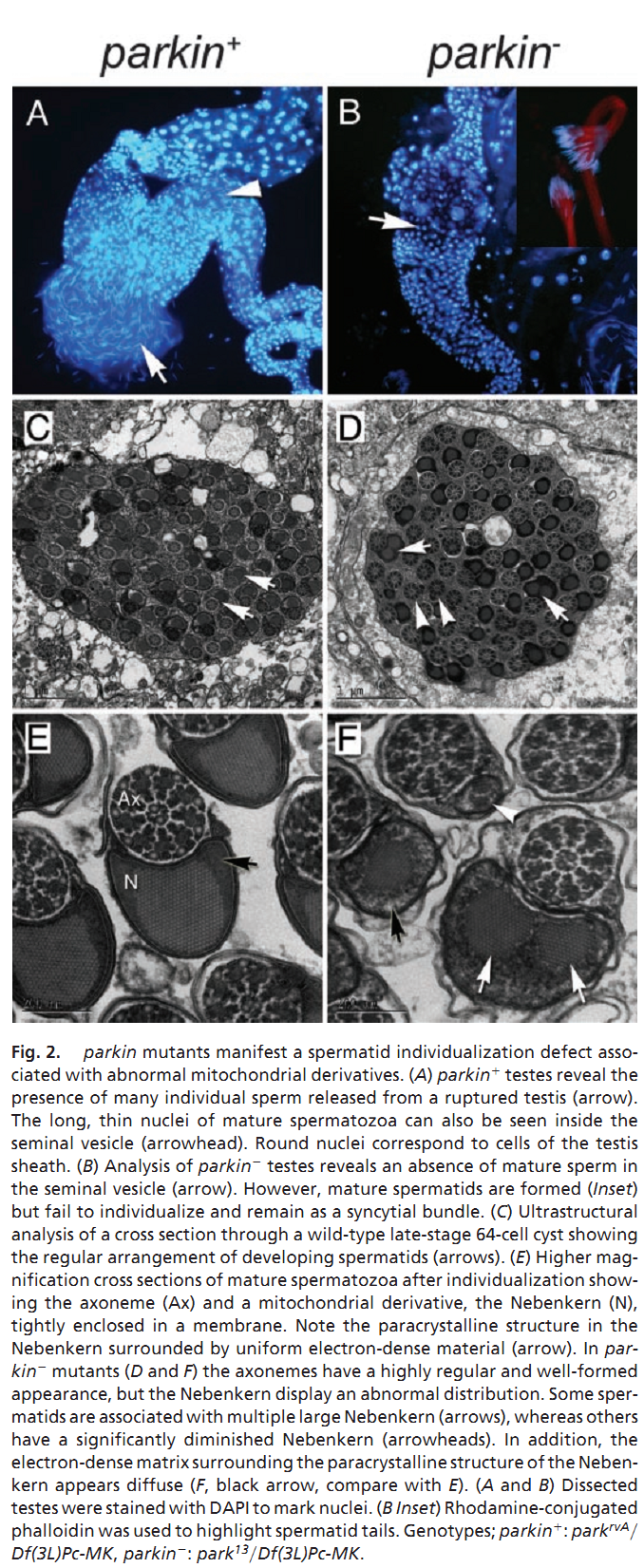
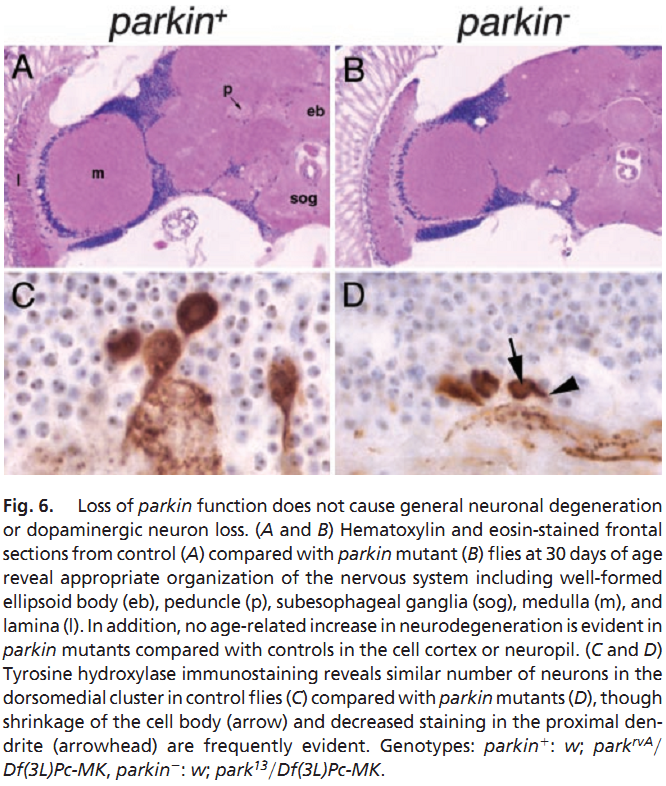
1. M. G. Riparbelli and G. Callaini (2007) The Drosophila parkin homologue is required for normal mitochondrial dynamics during spermiogenesis. Dev Biol 303(1): 108-20. Abstract Drosophila parkin, the ortholog of the human parkin gene, responsible for a familiar form of autosomal recessive juvenile parkinsonism, has been shown previously to be involved in Drosophila male fertility. Loss-of-function mutations in the parkin gene cause failure of spermatid individualization by affecting the proper progression of the actin-based investment cones that assemble in the nuclear region, but fail to translocate in synchrony down the cyst. In parkin mutants, the investment cones are scattered along the post-elongated spermatid bundles and fail to act properly in the process of sperm individualization. Using phase-contrast and electron microscopy analysis, we demonstrate that the parkin spermatids assemble a seemingly normal onion-stage nebenkern, but when the axoneme elongates only one mitochondrial derivative unfurls from the nebenkern. This unique mitochondrial derivative undergoes abnormal shaping and condensation during spermatid elongation. Our results indicate that parkin gene function is necessary for mitochondrial morphogenesis during earlier and later phases of spermiogenesis. The failure of cyst individualization may be due to the sensitivity of investment cone movement to the perturbation of mitochondrial morphology during spermatid elongation. PMID: [17123504] 2. J. C. Greene, A. J. Whitworth, I. Kuo, L. A. Andrews, M. B. Feany and L. J. Pallanck (2003) Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A 100(7): 4078-83. Abstract Parkinson's disease (PD) is a common neurodegenerative disorder characterized by loss of dopaminergic neurons in the substantia nigra. Several lines of evidence strongly implicate mitochondrial dysfunction as a major causative factor in PD, although the molecular mechanisms responsible for mitochondrial dysfunction are poorly understood. Recently, loss-of-function mutations in the parkin gene, which encodes a ubiquitin-protein ligase, were found to underlie a familial form of PD known as autosomal recessive juvenile parkinsonism (AR-JP). To gain insight into the molecular mechanism responsible for selective cell death in AR-JP, we have created a Drosophila model of this disorder. Drosophila parkin null mutants exhibit reduced lifespan, locomotor defects, and male sterility. The locomotor defects derive from apoptotic cell death of muscle subsets, whereas the male sterile phenotype derives from a spermatid individualization defect at a late stage of spermatogenesis. Mitochondrial pathology is the earliest manifestation of muscle degeneration and a prominent characteristic of individualizing spermatids in parkin mutants. These results indicate that the tissue-specific phenotypes observed in Drosophila parkin mutants result from mitochondrial dysfunction and raise the possibility that similar mitochondrial impairment triggers the selective cell loss observed in AR-JP. PMID: [12642658] Back to Top |
||||||||||||||||||||||||||||||||||||||||||
Figures for illustrating the function of this protein/gene |
|
||||||||||||||||||||||||||||||||||||||||||
Function |
|||||||||||||||||||||||||||||||||||||||||||
Subcellular Location |
|||||||||||||||||||||||||||||||||||||||||||
Tissue Specificity |
|||||||||||||||||||||||||||||||||||||||||||
Gene Ontology |
|
||||||||||||||||||||||||||||||||||||||||||
Interpro |
|||||||||||||||||||||||||||||||||||||||||||
Pfam |
|||||||||||||||||||||||||||||||||||||||||||
SMART |
|||||||||||||||||||||||||||||||||||||||||||
PROSITE |
|||||||||||||||||||||||||||||||||||||||||||
PRINTS |
|||||||||||||||||||||||||||||||||||||||||||
Created Date |
18-Oct-2012 |
||||||||||||||||||||||||||||||||||||||||||
Record Type |
Experiment identified |
||||||||||||||||||||||||||||||||||||||||||
Protein sequence Annotation |
|||||||||||||||||||||||||||||||||||||||||||
Nucleotide Sequence |
Length: 1478 bp Go to nucleotide: FASTA |
||||||||||||||||||||||||||||||||||||||||||
Protein Sequence |
Length: 482 bp Go to amino acid: FASTA |
||||||||||||||||||||||||||||||||||||||||||
The verified Protein-Protein interaction information |
|||||||||||||||||||||||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
||||||||||||||||||||||||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
||||||||||||||||||||||||||||||||||||||||||
Comments |
|||||||||||||||||||||||||||||||||||||||||||