| Tag | Content | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00000363 |
||||||||||||||||||||||||
UniProt Accession |
|||||||||||||||||||||||||
Theoretical PI |
6.34
|
||||||||||||||||||||||||
Molecular Weight |
145958 Da
|
||||||||||||||||||||||||
Genbank Nucleotide ID |
|||||||||||||||||||||||||
Genbank Protein ID |
|||||||||||||||||||||||||
Gene Name |
bru |
||||||||||||||||||||||||
Gene Synonyms/Alias |
ORFNames=CG2478 |
||||||||||||||||||||||||
Protein Name |
Protein brunelleschi |
||||||||||||||||||||||||
Protein Synonyms/Alias |
NIK- and IKBKB-binding protein; |
||||||||||||||||||||||||
Organism |
Drosophila melanogaster (Fruit fly) |
||||||||||||||||||||||||
NCBI Taxonomy ID |
7227 |
||||||||||||||||||||||||
Chromosome Location |
|
||||||||||||||||||||||||
Function in Stage |
|||||||||||||||||||||||||
Function in Cell Type |
|||||||||||||||||||||||||
Description |
Temporarily unavailable |
||||||||||||||||||||||||
The information of related literatures |
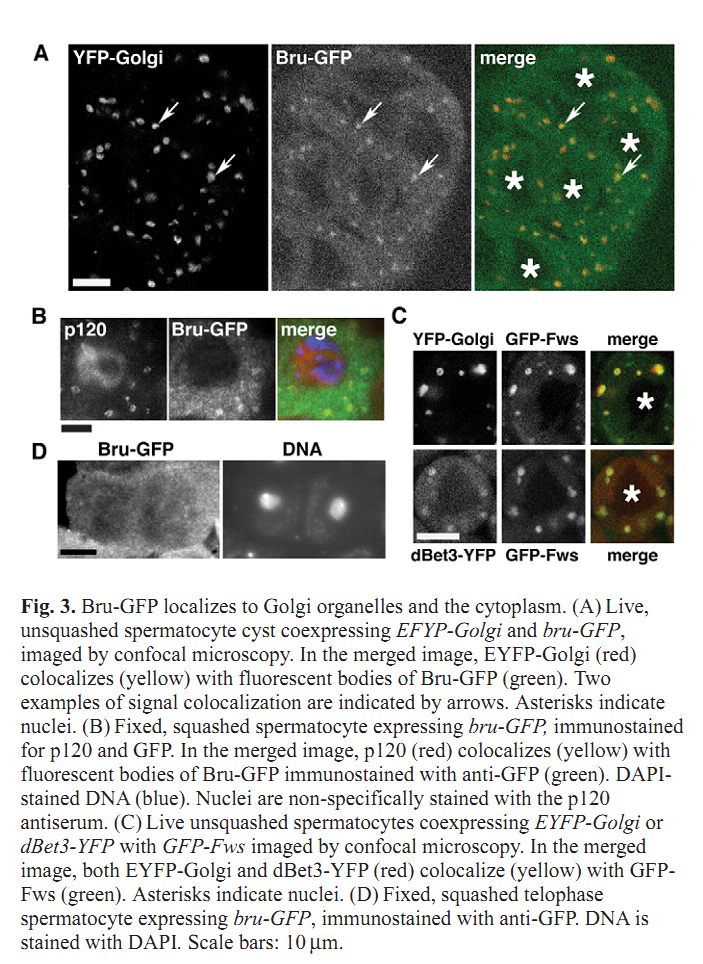
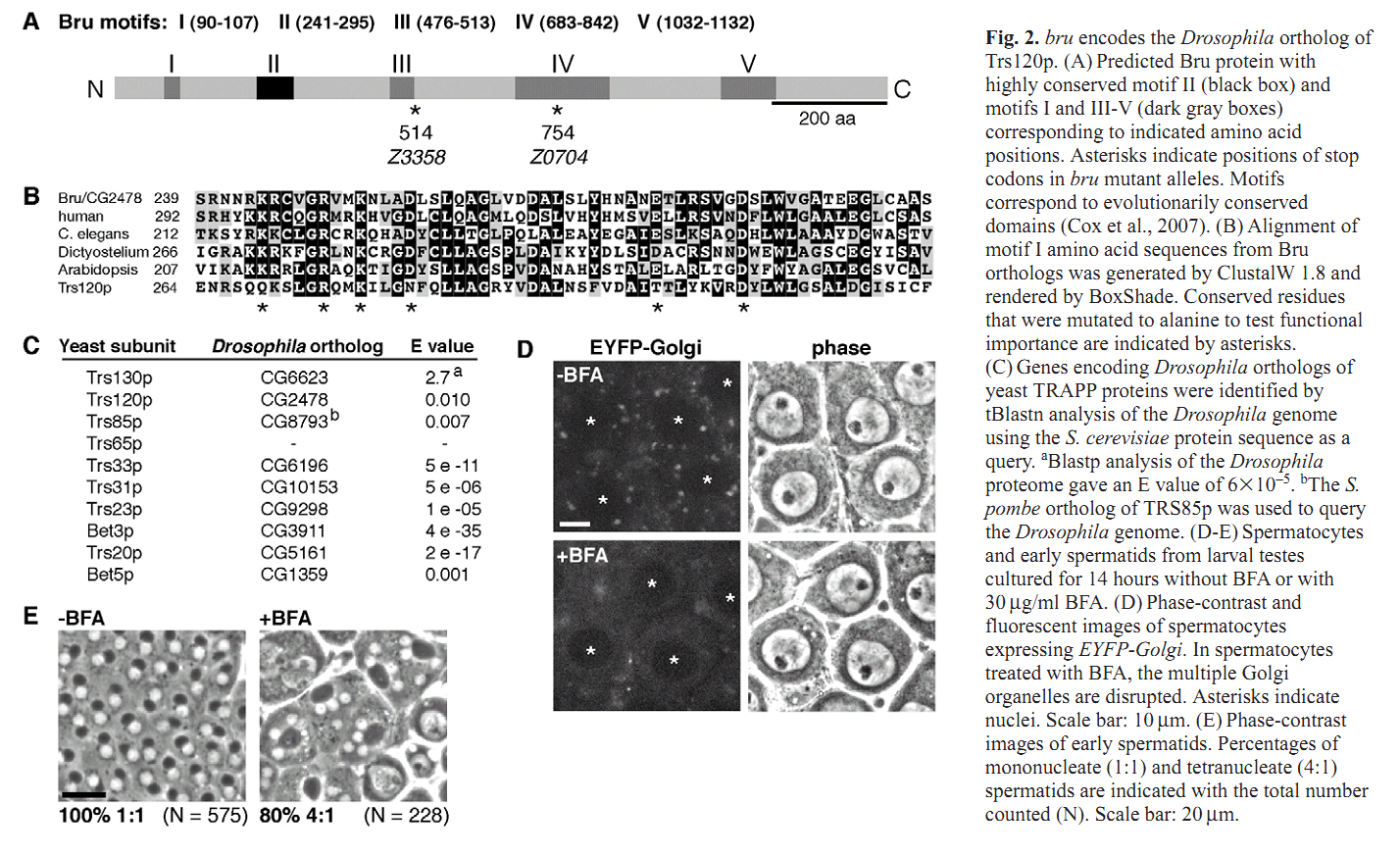
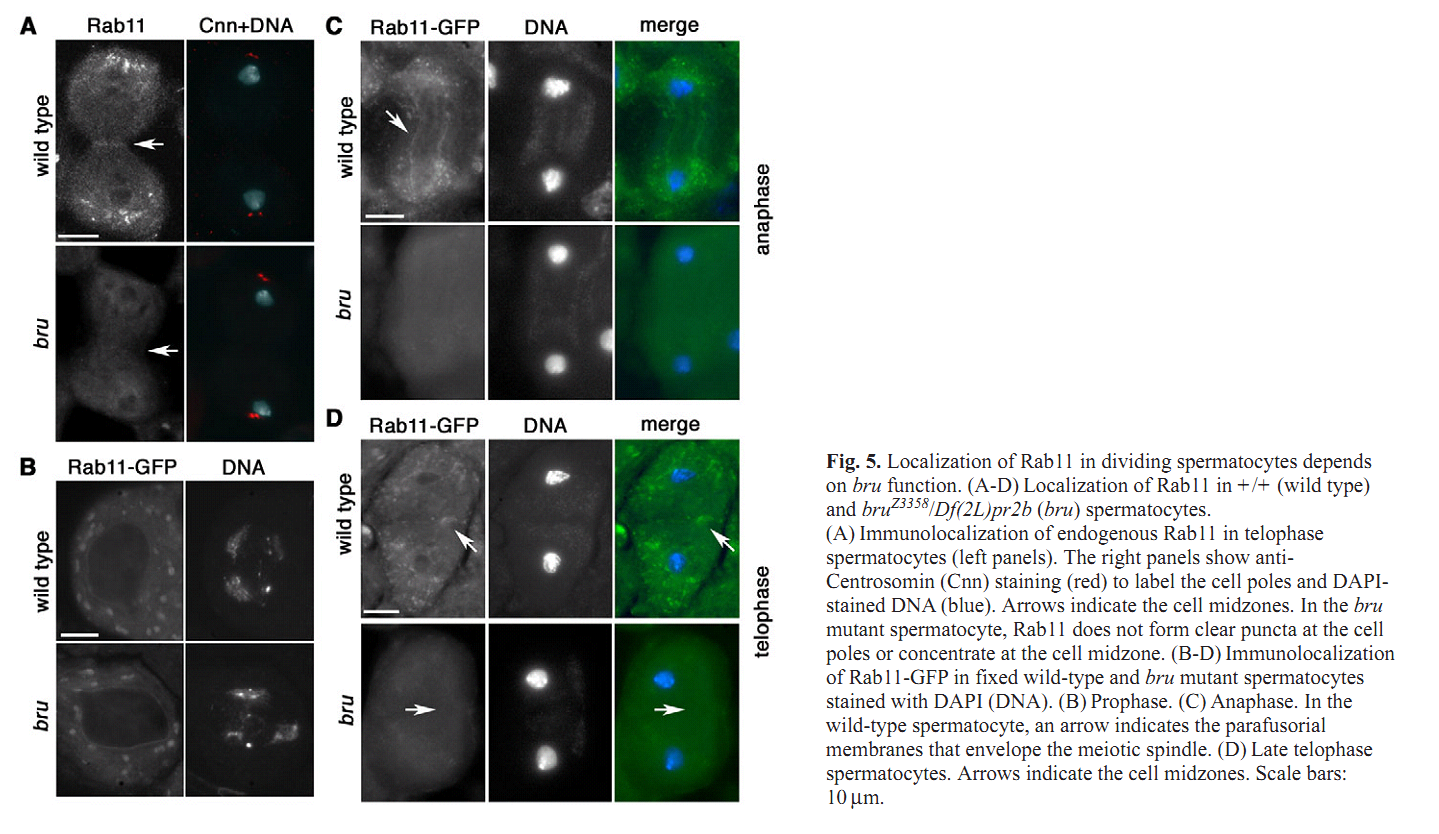
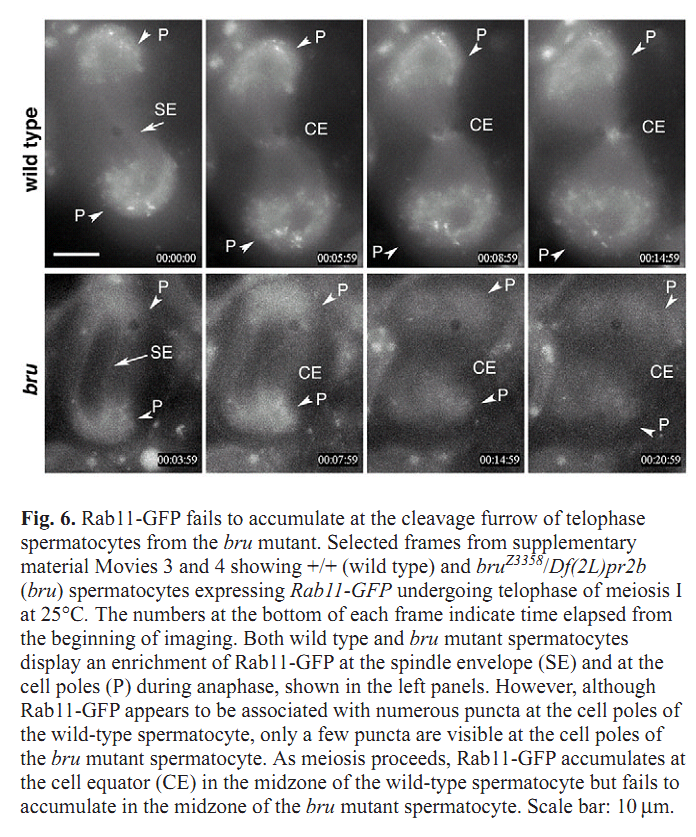
1. C. C. Robinett, M. G. Giansanti, M. Gatti and M. T. Fuller (2009) TRAPPII is required for cleavage furrow ingression and localization of Rab11 in dividing male meiotic cells of Drosophila. J Cell Sci 122(Pt 24): 4526-34. Abstract Although membrane addition is crucial for cytokinesis in many animal cell types, the specific mechanisms supporting cleavage furrow ingression are not yet understood. Mutations in the gene brunelleschi (bru), which encodes the Drosophila ortholog of the yeast Trs120p subunit of TRAPPII, cause failure of furrow ingression in male meiotic cells. In non-dividing cells, Brunelleschi protein fused to GFP is dispersed throughout the cytoplasm and enriched at Golgi organelles, similarly to another Drosophila TRAPPII subunit, dBet3. Localization of the membrane-trafficking GTPase Rab11 to the cleavage furrow requires wild-type function of bru, and genetic interactions between bru and Rab11 increase the failure of meiotic cytokinesis and cause synthetic lethality. bru also genetically interacts with four wheel drive (fwd), which encodes a PI4Kbeta, such that double mutants exhibit enhanced failure of male meiotic cytokinesis. These results suggest that Bru cooperates with Rab11 and PI4Kbeta to regulate the efficiency of membrane addition to the cleavage furrow, thus promoting cytokinesis in Drosophila male meiotic cells. PMID: [19934220] Back to Top |
||||||||||||||||||||||||
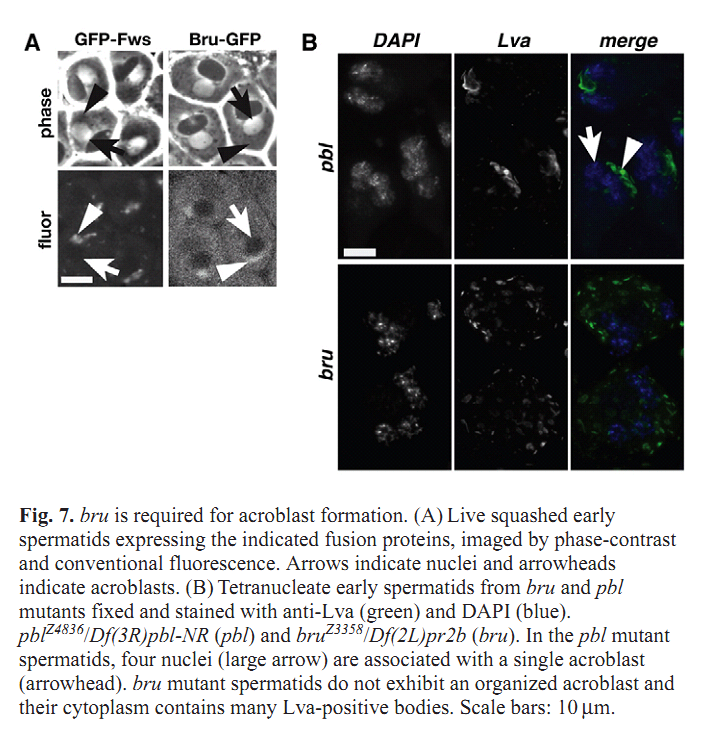
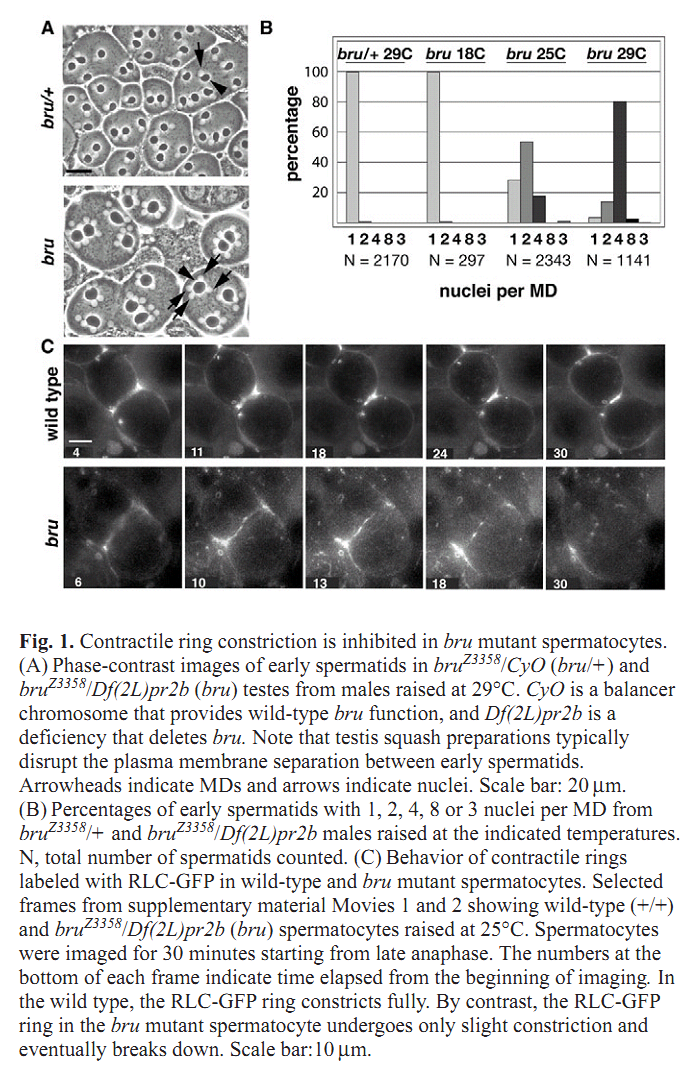
Figures for illustrating the function of this protein/gene |
|
||||||||||||||||||||||||
Function |
Cooperates with Rab11 and fwd/PI4K to mediate the flowof membrane through the Golgi, which is required to supportcleavage furrow ingression, therefore promoting cytokinesis inmale meiotic cells. Back to Top |
||||||||||||||||||||||||
Subcellular Location |
Cytoplasm. Golgi apparatus. Note=Specific tospermatocytes. |
||||||||||||||||||||||||
Tissue Specificity |
|||||||||||||||||||||||||
Gene Ontology |
|
||||||||||||||||||||||||
Interpro |
|||||||||||||||||||||||||
Pfam |
|||||||||||||||||||||||||
SMART |
|||||||||||||||||||||||||
PROSITE |
|||||||||||||||||||||||||
PRINTS |
|||||||||||||||||||||||||
Created Date |
18-Oct-2012 |
||||||||||||||||||||||||
Record Type |
Experiment identified |
||||||||||||||||||||||||
Protein sequence Annotation |
CHAIN 1 1320 Protein brunelleschi. /FTId=PRO_0000372863. COMPBIAS 363 398 Ser-rich. COMPBIAS 969 975 Poly-Gly. MOD_RES 317 317 Phosphoserine. MOD_RES 329 329 Phosphothreonine. MOD_RES 672 672 Phosphoserine. CONFLICT 83 83 H -> Y (in Ref. 3; ABA81812). CONFLICT 204 204 V -> I (in Ref. 3; ABA81812). CONFLICT 220 220 V -> M (in Ref. 3; ABA81812). CONFLICT 354 354 S -> C (in Ref. 3; ABA81812). Back to Top |
||||||||||||||||||||||||
Nucleotide Sequence |
Length: bp Go to nucleotide: FASTA |
||||||||||||||||||||||||
Protein Sequence |
Length: 1320 bp Go to amino acid: FASTA |
||||||||||||||||||||||||
The verified Protein-Protein interaction information |
|||||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
||||||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
||||||||||||||||||||||||
Comments |
|||||||||||||||||||||||||