| Tag | Content | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00000655 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
UniProt Accession |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Theoretical PI |
9.23
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Weight |
82764 Da
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genbank Nucleotide ID |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genbank Protein ID |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Name |
DAZ1 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Synonyms/Alias |
DAZ, SPGY |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Name |
Deleted in azoospermia protein 1 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Synonyms/Alias |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Organism |
Homo sapiens (Human) |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
NCBI Taxonomy ID |
9606 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Chromosome Location |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function in Stage |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function in Cell Type |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Description |
Larger deletions involving more than one AZF-candidate gene are associated with a more severe testicular phenotype. Partial deletion of the DAZ cluster could be cause of impaired spermatogenesis. DAZ gene activity seems to correspond to the proliferative activity of stem cells of germinal epithelium. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The information of related literatures |
1. R. Behulova, L. Strhakova, I. Boronova, A. Cibulkova, M. Konecny, L. Danisovic and V. Repiska (2011) DNA analysis of Y chromosomal AZF region in Slovak population with fertility disorders. Bratisl Lek Listy 112(4): 183-7. Abstract BACKGROUND PMID: [21585124] 2. A. Ferlin, E. Moro, A. Garolla and C. Foresta (1999) Human male infertility and Y chromosome deletions. Hum Reprod 14(7): 1710-6. Abstract Microdeletions in Yq11 overlapping three distinct 'azoospermia factors' (AZFa-c) represent the aetiological factor of 10-15% of idiopathic azoospermia and severe oligozoospermia, with higher prevalence in more severe testiculopathies, such as Sertoli cell-only syndrome. Using a PCR-based screening, we analysed Yq microdeletions in 180 infertile patients affected by idiopathic Sertoli cell-only syndrome and different degrees of hypospermatogenesis, compared with 50 patients with known causes of testicular alteration, 30 with obstructive azoospermia, and 100 normal fertile men. In idiopathic severe testiculopathies (Sertoli cell-only syndrome and severe hypospermatogenesis), a high prevalence of microdeletions (34.5% and 24.7% respectively) was found, while milder forms were not associated with Yq alteration. No deletions were found in testiculopathies of known aetiology, obstructive azoospermia, normal fertile men and male relatives of patients with deletions. Deletions in the AZFc region involving the DAZ gene were the most frequent finding and they were more often observed in severe hypospermatogenesis than in Sertoli cell-only syndrome, suggesting that deletions of this region are not sufficient to cause complete loss of the spermatogenic line. Deletions in AZFb involving the RBM gene were less frequently detected and there was no correlation with testicular phenotype, with an apparent minor role for such gene in spermatogenesis. The DFFRY gene was absent in a fraction of patients, making it a candidate AZFa gene. Our data suggest that larger deletions involving more than one AZF-candidate gene are associated with a more severe testicular phenotype. PMID: [10402373] 3. T. Bienvenu, C. Patrat, K. McElreavey, M. de Almeida and P. Jouannet (2001) Reduction in the DAZ gene copy number in two infertile men with impaired spermatogenesis. Ann Genet 44(3): 125-8. Abstract Microdeletions of Yq are associated with azoospermia and severe oligozoospermia. Recently, we described a new molecular genetic strategy based on the denaturing gradient gel electrophoresis (DGGE) to rapidly identify deletions of the Y chromosome that include the DAZ locus. Using this approach, we have shown not a complete deletion but only a reduction in the number of copies of the DAZ gene by PCR-DGGE in two oligozoospermic patients. These results have been confirmed by Southern blot analysis. This finding suggests that partial deletion of the DAZ cluster could be cause of impaired spermatogenesis. PMID: [11694223] 4. S. Fernandes, K. Huellen, J. Goncalves, H. Dukal, J. Zeisler, E. Rajpert De Meyts, N. E. Skakkebaek, B. Habermann, W. Krause, M. Sousa, A. Barros and P. H. Vogt (2002) High frequency of DAZ1/DAZ2 gene deletions in patients with severe oligozoospermia. Mol Hum Reprod 8(3): 286-98. Abstract Deletions of the DAZ gene family in distal Yq11 are always associated with deletions of the azoospermia factor c (AZFc) region, which we now estimate extends to 4.94 Mb. Because more Y gene families are located in this chromosomal region, and are expressed like the DAZ gene family only in the male germ line, the testicular pathology associated with complete AZFc deletions cannot predict the functional contribution of the DAZ gene family to human spermatogenesis. We therefore established a DAZ gene copy specific deletion analysis based on the DAZ-BAC sequences in GenBank. It includes the deletion analysis of eight DAZ-DNA PCR markers [six DAZ-single nucleotide varients (SNVs) and two DAZ-sequence tag sites (STS)] selected from the 5' to the 3'end of each DAZ gene and a deletion analysis of the gene copy specific EcoRV and TaqI restriction fragments identified in the internal repetitive DAZ gene regions (DYS1 locus). With these diagnostic tools, 63 DNA samples from men with idiopathic oligozoospermia and 107 DNA samples from men with proven fertility were analysed for the presence of the complete DAZ gene locus, encompassing the four DAZ gene copies. In five oligozoospermic patients, we found a DAZ-SNV/STS and DYS1/EcoRV and TaqI fragment deletion pattern indicative for deletion of the DAZ1 and DAZ2 gene copies; one of these deletions could be identified as a 'de-novo' deletion because it was absent in the DAZ locus of the patient's father. The same DAZ deletions were not found in any of the 107 fertile control samples. We therefore conclude that the deletion of the DAZ1/DAZ2 gene doublet in five out of our 63 oligozoospermic patients (8%) is responsible for the patients' reduced sperm numbers. It is most likely caused by intrachromosomal recombination events between two long repetitive sequence blocks (AZFc-Rep1) flanking the DAZ gene structures. PMID: [11870237] 5. A. Ferlin, A. Tessari, F. Ganz, E. Marchina, S. Barlati, A. Garolla, B. Engl and C. Foresta (2005) Association of partial AZFc region deletions with spermatogenic impairment and male infertility. J Med Genet 42(3): 209-13. Abstract BACKGROUND PMID: [15744033] 6. W. J. Huang, Y. W. Lin, K. N. Hsiao, K. S. Eilber, E. C. Salido and P. H. Yen (2008) Restricted expression of the human DAZ protein in premeiotic germ cells. Hum Reprod 23(6): 1280-9. Abstract BACKGROUND PMID: [18385127] 7. P. L. Kuo, S. T. Wang, Y. M. Lin, Y. H. Lin, Y. N. Teng and C. C. Hsu (2004) Expression profiles of the DAZ gene family in human testis with and without spermatogenic failure. Fertil Steril 81(4): 1034-40. Abstract OBJECTIVE PMID: [15066460] 8. L. Becherini, E. Guarducci, S. Degl'Innocenti, M. Rotondi, G. Forti and C. Krausz (2004) DAZL polymorphisms and susceptibility to spermatogenic failure. Int J Androl 27(6): 375-81. Abstract Polymorphisms in genes involved in spermatogenesis are considered potential risk factors for male infertility. Recently a polymorphism in the deleted in azoospermia-like (DAZL) gene (T54A) was reported as susceptibility factor to oligo/azoospermia in the Chinese population. DAZL is an autosomal homologue of the Y chromosomal DAZ (deleted in azoospermia) gene cluster and both are considered master regulators of spermatogenesis. The aim of the present study was to screen (i) for mutations of the entire coding sequence of the DAZL gene in patients lacking of the DAZ gene cluster, in order to evaluate if DAZL polymorphisms may influence the AZFc deletion phenotype; (ii) for the two previously described (and eventually newly identified) single nucleotide polymorphisms (SNPs) in a large group of infertile and normospermic men of Italian origin. We failed to detect new mutations. We confirmed previous results showing no evidence for a functional role of the T12A mutation. Surprisingly, the T54A polymorphism, which was present in 7.4% of the Chinese patients was absent in our Caucasian population. This remarkable difference represent an example of how ethnic background is important also for polymorphisms involved in spermatogenesis and contributes to better select clinically relevant tests, specifically based on the ethnic origin of the infertile patients. PMID: [15595957] 9. C. Ferras, S. Fernandes, C. J. Marques, F. Carvalho, C. Alves, J. Silva, M. Sousa and A. Barros (2004) AZF and DAZ gene copy-specific deletion analysis in maturation arrest and Sertoli cell-only syndrome. Mol Hum Reprod 10(10): 755-61. Abstract Deletions of the AZFc region in Yq11.2, which include the DAZ gene family, are responsible for most cases of male infertility and were associated with severe oligozoospermia and also with a variable testicular pathology. To uncover the functional contribution of DAZ to human spermatogenesis, a DAZ gene copy-specific deletion analysis was previously established and showed that DAZ1/DAZ2 deletions associate with oligozoospermia. In this study we applied the same screening method to 50 control fertile males and 91 non-obstructive azoospermic males, 39 with Sertoli cell-only syndrome (SCOS) and 52 with meiotic arrest (MA). Samples were also screened with 24 sequence-tagged sites to the different AZF regions, including 114 control fertile males. After biopsy (testicular sperm extraction, TESE), residual spermiogenesis was found in 57.7% MA and 30.8% SCOS cases (incomplete syndromes). DAZ1/DAZ2 deletions were associated with the testicular phenotype of residual spermiogenesis as they were only found in two patients (8%) with incomplete MA. Differences between incomplete (23.3%) and complete (4.5%) MA cases regarding AZFc and DAZ1/DAZ2 deletion frequencies, and between incomplete (58.3%) and complete (11.1%) SCOS cases for AZFc deletions, suggest that incomplete syndromes might represent an aggravation of the oligozoospermic phenotype. As successful TESE was achieved in 87.5% of MA cases with AZFc and DAZ1/DAZ2 deletions and in 58.3% of SCOS cases with AZFc deletions, the present results also suggest that these molecular markers might be used for the establishment of a prognosis before TESE. PMID: [15347736] 10. A. Szczerba, A. Jankowska, M. Andrusiewicz and J. B. Warchol (2006) Localization of the DAZ gene expression in seminiferous tubules of patients with spermatogenic disorders. Folia Histochem Cytobiol 44(2): 123-6. Abstract The research on the expression and mutations of DAZ and its homologues in human and other mammals suggests that protein products of these genes can mainly affect development of germinal cells. The aim of the present study was to analyze the expression of the DAZ gene in seminiferous tubules of six men with spermatogenic disorders (hypospermatogenesis and spermatogenic arrest). The results based on the RT-PCR IS technique demonstrated that the DAZ product was present only in some seminiferous tubules and the fluorescence intensity was different within individual tubules. The most intense fluorescence characterised the spermatogonia, especially these organised in small groups inside separate tubules. In the patients with spermatogenic arrest at the spermatocyte stage, the DAZ gene transcripts were also found in primary spermatocytes. However, the fluorescence intensity of primary spermatocytes, except the fluorescence of the spermatocytes localised upon the lumen, was weaker than the fluorescence of spermatogonia. The results of our study showed that DAZ gene activity seems to correspond to the proliferative activity of stem cells of germinal epithelium. PMID: [16805138] 11. J. Gianotten, M. J. Hoffer, J. W. De Vries, N. J. Leschot, J. Gerris and F. van der Veen (2003) Partial DAZ deletions in a family with five infertile brothers. Fertil Steril 79 Suppl 3(): 1652-5. Abstract OBJECTIVE PMID: [12801575] 12. Y. Yang, C. Xiao, S. Zhang, A. Zhoucun and X. Li (2006) Preliminary study of the relationship between DAZ gene copy deletions and spermatogenic impairment in Chinese men. Fertil Steril 85(4): 1061-3. Abstract Copy deletion screening of DAZ gene family on the Y chromosome in 485 patients with idiopathic azoospermia or oligozoospermia and 236 fertile men revealed that the prevalence of deletion patterns of the entire DAZ gene and DAZ1/DAZ2 gene were significantly higher in the patients than in fertile men. The deletion patterns correlate with spermatogenic impairment in a Chinese population. PMID: [16580401] 13. A. Ferlin, E. Moro, M. Onisto, E. Toscano, A. Bettella and C. Foresta (1999) Absence of testicular DAZ gene expression in idiopathic severe testiculopathies. Hum Reprod 14(9): 2286-92. Abstract Deletions of the DAZ (deleted in azoospermia) gene family are frequently responsible for male infertility and are generally assessed by analyses of genomic DNA extracted from peripheral leukocytes. The multicopy nature of this gene prevents the distinction of intragenic deletions or deletions not involving the whole DAZ gene cluster. Thus it is still unclear whether each DAZ copy is effectively expressed in the testis. We analysed, by reverse transcription-polymerase chain reaction (RT-PCR), the expression of DAZ, RBM and SRY genes, in testicular cells from infertile men affected by idiopathic severe hypospermatogenesis, obstructive azoospermia and Sertoli cell-only syndrome. Normal mRNA for DAZ, RBM and SRY were observed in obstructive azoospermia, whereas only SRY transcripts were detected when only Sertoli cells were present. Nine out of 10 patients affected by idiopathic severe hypospermatogenesis had normal expression of SRY, RBM and DAZ, while in one patient no DAZ transcript was detected, suggesting that his testiculopathy was related to the absence of DAZ expression. The lack of DAZ mRNA in testicular cells with an apparently normal DAZ gene constitution on DNA extracted from leukocytes may be explained by different hypotheses PMID: [10469697] 14. A. Szczerba, A. Jankowska, M. Andrusiewicz, M. Karczewski, W. Turkiewicz and J. B. Warchol (2004) Distribution of the DAZ gene transcripts in human testis. Folia Histochem Cytobiol 42(2): 119-21. Abstract Involvement of variety of genes, especially located on Y chromosome, is critical for the regulation of spermatogenesis. In particular, fertility candidate genes such as deleted in azoospermia (DAZ) are believed to have important function in sperm production, since DAZ is frequently deleted in azoospermic and severy oligozoospermic men. The role of the DAZ gene is supported by its exclusive expression in the testis and by its deletion in about 10% of azoospermic and severely oligozoospermic patients. The distribution of DAZ transcripts in seminiferous epithelium of human testis is reported in the present study. The use of Adobe Photoshop and Scion Image softwares allowed for semi-quantitative analysis of in situ RT-PCR (ISRT-PCR) results. The intensity of ISRT-PCR product's fluorescence was different within individual seminiferous tubules. It was clearly shown by using the pseudocolour scale and transforming the intensity of the fluorescence into levels of greyscale images. The more intense fluorescence characterised single spermatogonia and those organized in small groups inside separate tubules. The most intense accumulation of DAZ mRNA was observed in spermatogonia. PMID: [15253135] Back to Top |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
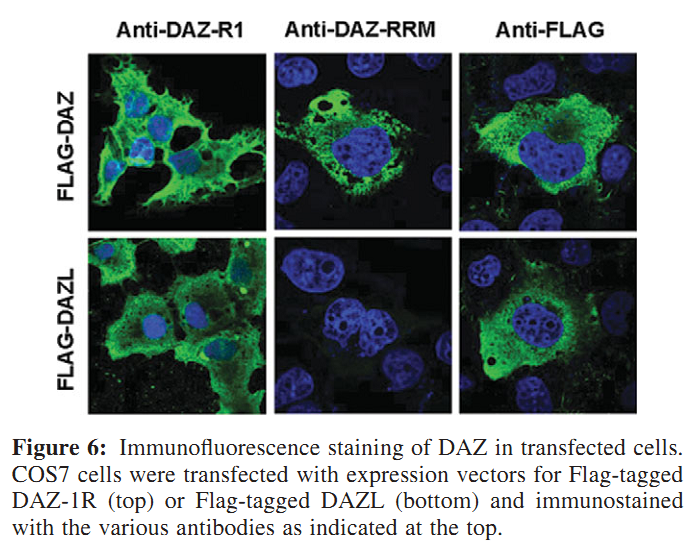
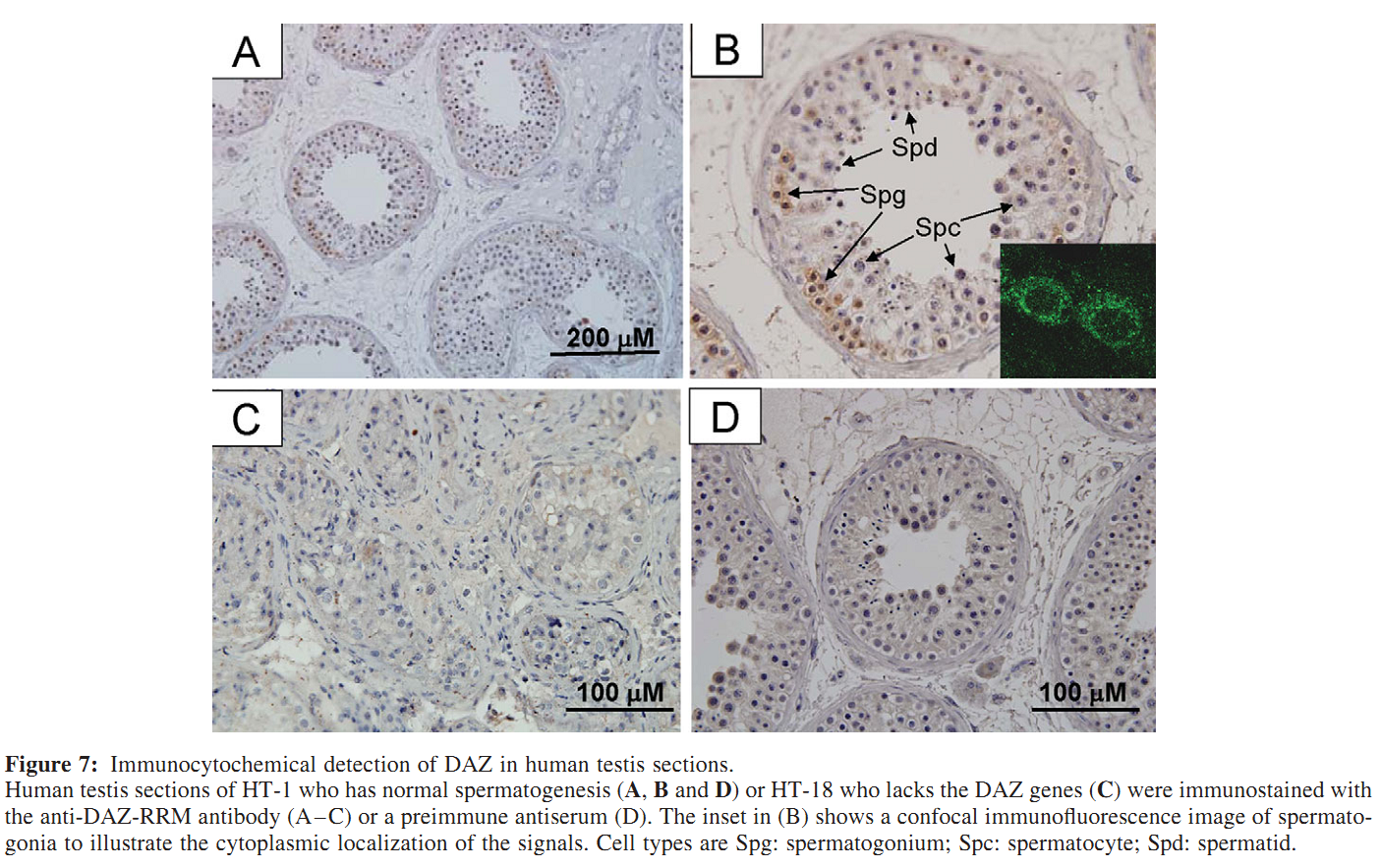
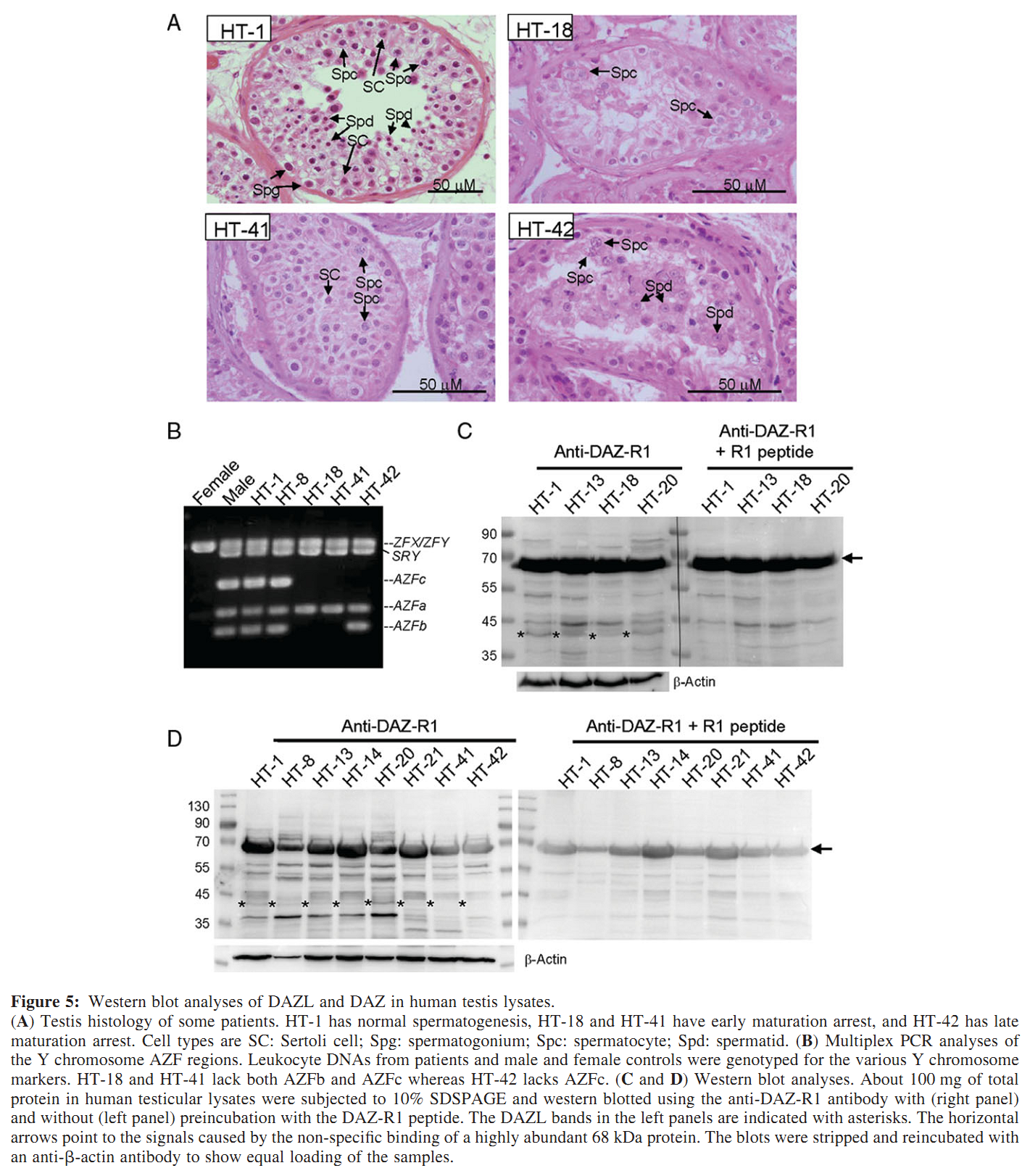
Figures for illustrating the function of this protein/gene |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function |
RNA-binding protein that plays an essential role inspermatogenesis. May act by binding to the 3'-UTR of mRNAs andregulating their translation. Promotes germ-cell progression tomeiosis and formation of haploid germ cells. Back to Top |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subcellular Location |
Cytoplasm. Nucleus. Note=Predominantlycytoplasmic. Nuclear at some stages of spermatozoide development.Localizes both to the nuclei and cytoplasm of spermatozoidedifferentiation. Nuclear in fetal gonocytes and in spermatogonialnuclei. It then relocates to the cytoplasm during male meiosis. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tissue Specificity |
Testis-specific. Expression restricted topremeiotic germ cells, particularly in spermatogonia (at proteinlevel). |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Ontology |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Interpro |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Pfam |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
SMART |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PROSITE |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PRINTS |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Created Date |
18-Oct-2012 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Record Type |
Experiment identified |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein sequence Annotation |
CHAIN 1 744 Deleted in azoospermia protein 1. /FTId=PRO_0000081554. DOMAIN 40 115 RRM 1. DOMAIN 205 280 RRM 2. DOMAIN 370 445 RRM 3. DOMAIN 500 520 DAZ-like 1. DOMAIN 524 544 DAZ-like 2. DOMAIN 549 569 DAZ-like 3. DOMAIN 572 592 DAZ-like 4. DOMAIN 596 616 DAZ-like 5. DOMAIN 620 640 DAZ-like 6. DOMAIN 644 664 DAZ-like 7. DOMAIN 668 688 DAZ-like 8. DOMAIN 692 712 DAZ-like 9. CONFLICT 287 287 R -> G (in Ref. 2; AAF91405). Back to Top |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Nucleotide Sequence |
Length: 175990 bp Go to nucleotide: FASTA |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Sequence |
Length: 744 bp Go to amino acid: FASTA |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The verified Protein-Protein interaction information |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Comments |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||