| Tag | Content | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00000816 |
|||||||||||||||||||||||||||||||||||||||||||||||||||
UniProt Accession |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Theoretical PI |
6.17
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Weight |
33751 Da
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
Genbank Nucleotide ID |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Genbank Protein ID |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Name |
Cdk4 |
|||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Synonyms/Alias |
Crk3 |
|||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Name |
Cyclin-dependent kinase 4 |
|||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Synonyms/Alias |
EC=2.7.11.22 CRK3; Cell division protein kinase 4; PSK-J3; |
|||||||||||||||||||||||||||||||||||||||||||||||||||
Organism |
Mus musculus (Mouse) |
|||||||||||||||||||||||||||||||||||||||||||||||||||
NCBI Taxonomy ID |
10090 |
|||||||||||||||||||||||||||||||||||||||||||||||||||
Chromosome Location |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
Function in Stage |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Function in Cell Type |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Description |
Cyclin-dependent kinase 4 (Cdk4) and Cdk6, and later Cdk2, in association with their specific cyclin partners, regulate the G1 to S phase cell cycle transition of mammalian cells by phosphorylation of retinoblastoma (Rb) family proteins. Loss of Cdk4 leads to an age-dependent defect in spermatogenesis and disruption in the timing of the estrus cycle.Cdk4 participates in proliferation of beta-islet cells. |
|||||||||||||||||||||||||||||||||||||||||||||||||||
The information of related literatures |
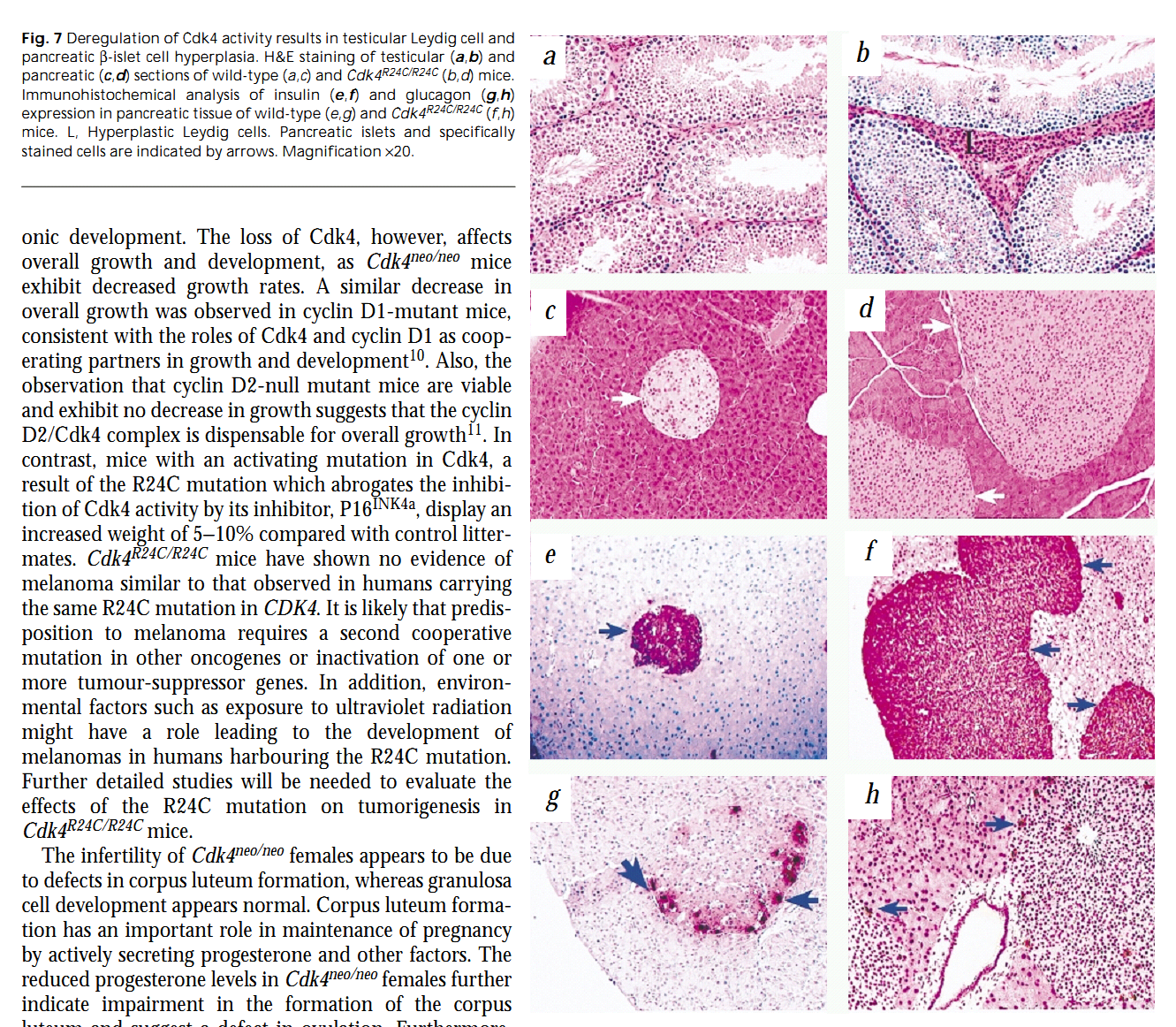
1. R. V. Mettus and S. G. Rane (2003) Characterization of the abnormal pancreatic development, reduced growth and infertility in Cdk4 mutant mice. Oncogene 22(52): 8413-21. Abstract Cyclin-dependent kinase 4 (Cdk4) and Cdk6, and later Cdk2, in association with their specific cyclin partners, regulate the G1 to S phase cell cycle transition of mammalian cells by phosphorylation of retinoblastoma (Rb) family proteins. Phosphorylation of Rb results in the release of S-phase specific transcription factors; cell cycle-promoting gene expression, and advancement of the cell cycle. Loss of Cdk4 by homologous-targeted disruption leads to a delay in S-phase entry in serum-stimulated mouse embryo fibroblast (MEF) cultures. Homozygous Cdk4-deficient mice display defects in weight gain, fertility and hypoproliferation of specific endocrine cells of the pituitary and pancreas, the latter of which results in a diabetes-like phenotype. In contrast, inheritance of the p16(Ink4a)-insensitive Cdk4(R24C) mutation leads to spontaneous transformation of MEF cultures in vitro and, in vivo, hyperproliferative disorders that progress to cancer. In this manuscript, we report characterization of the abnormal pancreatic development, reduced growth and infertility in Cdk4 mutant mice. We observe that, whereas Cdk4 is dispensable for early pancreatic development, normal Cdk4 expression is critical for optimal growth of the organism. Also, we observe that loss of Cdk4 may result in insulin insensitivity, implicating an additional role of Cdk4 in beta-cell function, in addition to its role in beta-cell proliferation. Further, we demonstrate that loss of Cdk4 leads to an age-dependent defect in spermatogenesis and disruption in the timing of the estrus cycle. Taken together, our results indicate that the overall defects in growth, fertility and pancreatic development in Cdk4-deficient mice may be a combination of cell-type specific defects and altered glucose metabolism, as a result of defects in postnatal pancreatic development. PMID: [14627982] 2. S. G. Rane, P. Dubus, R. V. Mettus, E. J. Galbreath, G. Boden, E. P. Reddy and M. Barbacid (1999) Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in beta-islet cell hyperplasia. Nat Genet 22(1): 44-52. Abstract To ascertain the role of cyclin-dependent kinase 4 (Cdk4) in vivo, we have targeted the mouse Cdk4 locus by homologous recombination to generate two strains of mice, one that lacks Cdk4 expression and one that expresses a Cdk4 molecule with an activating mutation. Embryonic fibroblasts proliferate normally in the absence of Cdk4 but have a delayed S phase on re-entry into the cell cycle. Moreover, mice devoid of Cdk4 are viable, but small in size and infertile. These mice also develop insulin-deficient diabetes due to a reduction in beta-islet pancreatic cells. In contrast, mice expressing a mutant Cdk4 that cannot bind the cell-cycle inhibitor P16INK4a display pancreatic hyperplasia due to abnormal proliferation of beta-islet cells. These results establish Cdk4 as an essential regulator of specific cell types. PMID: [10319860] Back to Top |
|||||||||||||||||||||||||||||||||||||||||||||||||||
Figures for illustrating the function of this protein/gene |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
Function |
Ser/Thr-kinase component of cyclin D-CDK4 (DC) complexesthat phosphorylate and inhibit members of the retinoblastoma (RB)protein family including RB1 and regulate the cell-cycle duringG(1)/S transition. Phosphorylation of RB1 allows dissociation ofthe transcription factor E2F from the RB/E2F complexes and thesubsequent transcription of E2F target genes which are responsiblefor the progression through the G(1) phase. Hypophosphorylates RB1in early G(1) phase. Cyclin D-CDK4 complexes are major integratorsof various mitogenenic and antimitogenic signals. Alsophosphorylates SMAD3 in a cell-cycle-dependent manner andrepresses its transcriptional activity. Component of the ternarycomplex, cyclin D/CDK4/CDKN1B, required for nuclear translocationand activity of the cyclin D-CDK4 complex (By similarity). Back to Top |
|||||||||||||||||||||||||||||||||||||||||||||||||||
Subcellular Location |
Cytoplasm (By similarity). Nucleus (Bysimilarity). Membrane (By similarity). Note=Cytoplasmic when non-complexed. Forms a cyclin D-CDK4 complex in the cytoplasm as cellsprogress through G(1) phase. The complex accumulates on thenuclear membrane and enters the nucleus on transition from G(1) toS phase. Also present in nucleoli and heterochromatin lumps.Colocalizes with RB1 after release into the nucleus (Bysimilarity). |
|||||||||||||||||||||||||||||||||||||||||||||||||||
Tissue Specificity |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Ontology |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
Interpro |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Pfam |
||||||||||||||||||||||||||||||||||||||||||||||||||||
SMART |
||||||||||||||||||||||||||||||||||||||||||||||||||||
PROSITE |
PS00107; PROTEIN_KINASE_ATP; 1. PS50011; PROTEIN_KINASE_DOM; 1. PS00108; PROTEIN_KINASE_ST; 1. Back to Top |
|||||||||||||||||||||||||||||||||||||||||||||||||||
PRINTS |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Created Date |
18-Oct-2012 |
|||||||||||||||||||||||||||||||||||||||||||||||||||
Record Type |
Experiment identified |
|||||||||||||||||||||||||||||||||||||||||||||||||||
Protein sequence Annotation |
INIT_MET 1 1 Removed (By similarity). CHAIN 2 303 Cyclin-dependent kinase 4. /FTId=PRO_0000085779. DOMAIN 6 295 Protein kinase. NP_BIND 12 20 ATP (By similarity). REGION 50 56 Required for binding D-type cyclins (By similarity). ACT_SITE 140 140 Proton acceptor (By similarity). BINDING 35 35 ATP (By similarity). MOD_RES 2 2 N-acetylalanine (By similarity). MOD_RES 150 150 Phosphoserine (By similarity). MOD_RES 172 172 Phosphothreonine; by CAK. Back to Top |
|||||||||||||||||||||||||||||||||||||||||||||||||||
Nucleotide Sequence |
Length: 912 bp Go to nucleotide: FASTA |
|||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Sequence |
Length: 303 bp Go to amino acid: FASTA |
|||||||||||||||||||||||||||||||||||||||||||||||||||
The verified Protein-Protein interaction information |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
|||||||||||||||||||||||||||||||||||||||||||||||||||
View Microarray data |
||||||||||||||||||||||||||||||||||||||||||||||||||||
Comments |
||||||||||||||||||||||||||||||||||||||||||||||||||||