| Tag | Content | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00001976 |
||||||||||||||||||||||||
UniProt Accession |
|||||||||||||||||||||||||
Theoretical PI |
6.91
|
||||||||||||||||||||||||
Molecular Weight |
95440 Da
|
||||||||||||||||||||||||
Genbank Nucleotide ID |
|||||||||||||||||||||||||
Genbank Protein ID |
|||||||||||||||||||||||||
Gene Name |
Odf2 |
||||||||||||||||||||||||
Gene Synonyms/Alias |
Odf84 |
||||||||||||||||||||||||
Protein Name |
Outer dense fiber protein 2 |
||||||||||||||||||||||||
Protein Synonyms/Alias |
84 kDa outer dense fiber protein; Cenexin; Outer dense fiber of sperm tails protein 2; |
||||||||||||||||||||||||
Organism |
Rattus norvegicus (Rat) |
||||||||||||||||||||||||
NCBI Taxonomy ID |
10116 |
||||||||||||||||||||||||
Chromosome Location |
|
||||||||||||||||||||||||
Function in Stage |
|||||||||||||||||||||||||
Function in Cell Type |
|||||||||||||||||||||||||
Description |
Temporarily unavailable |
||||||||||||||||||||||||
The information of related literatures |
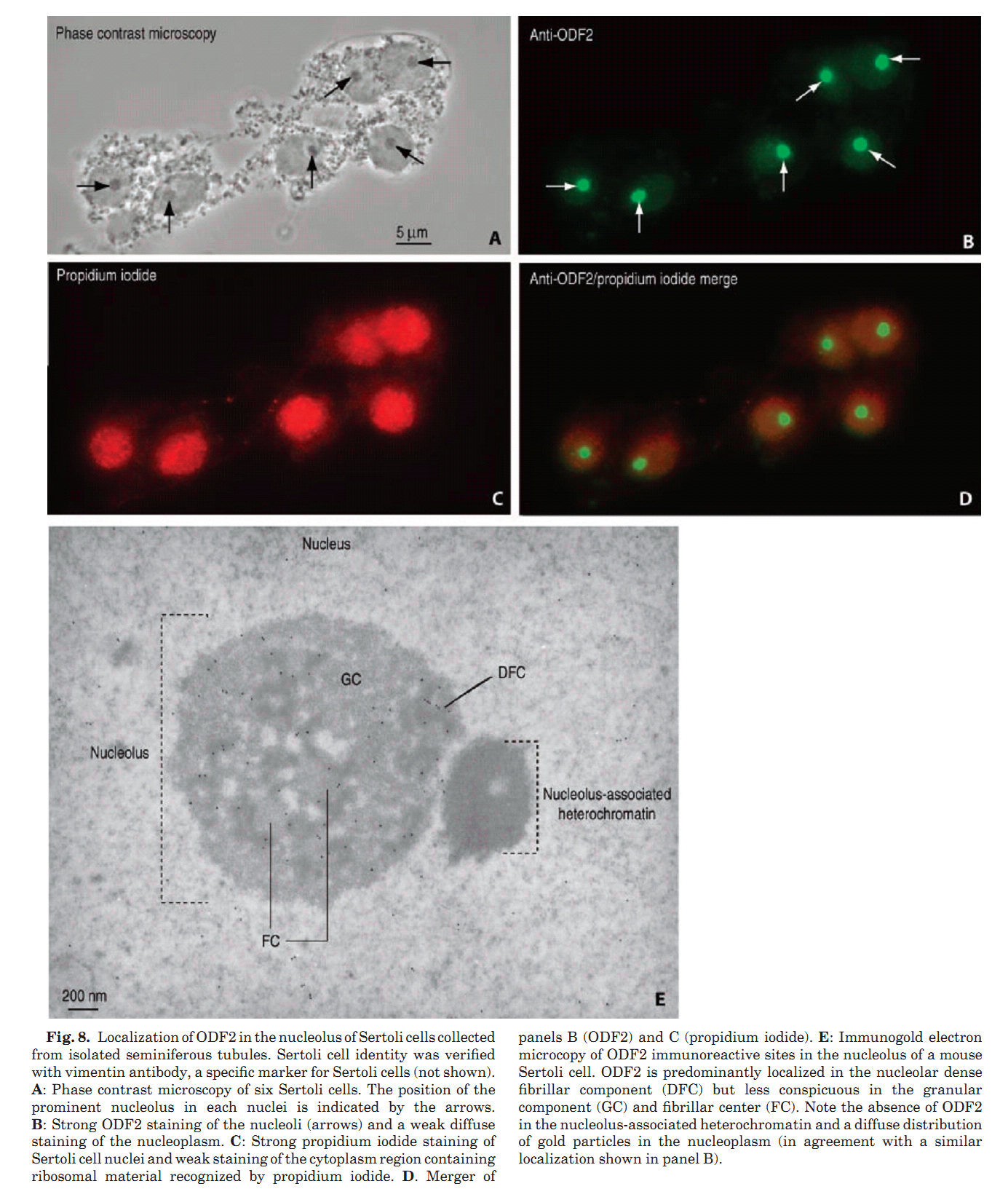
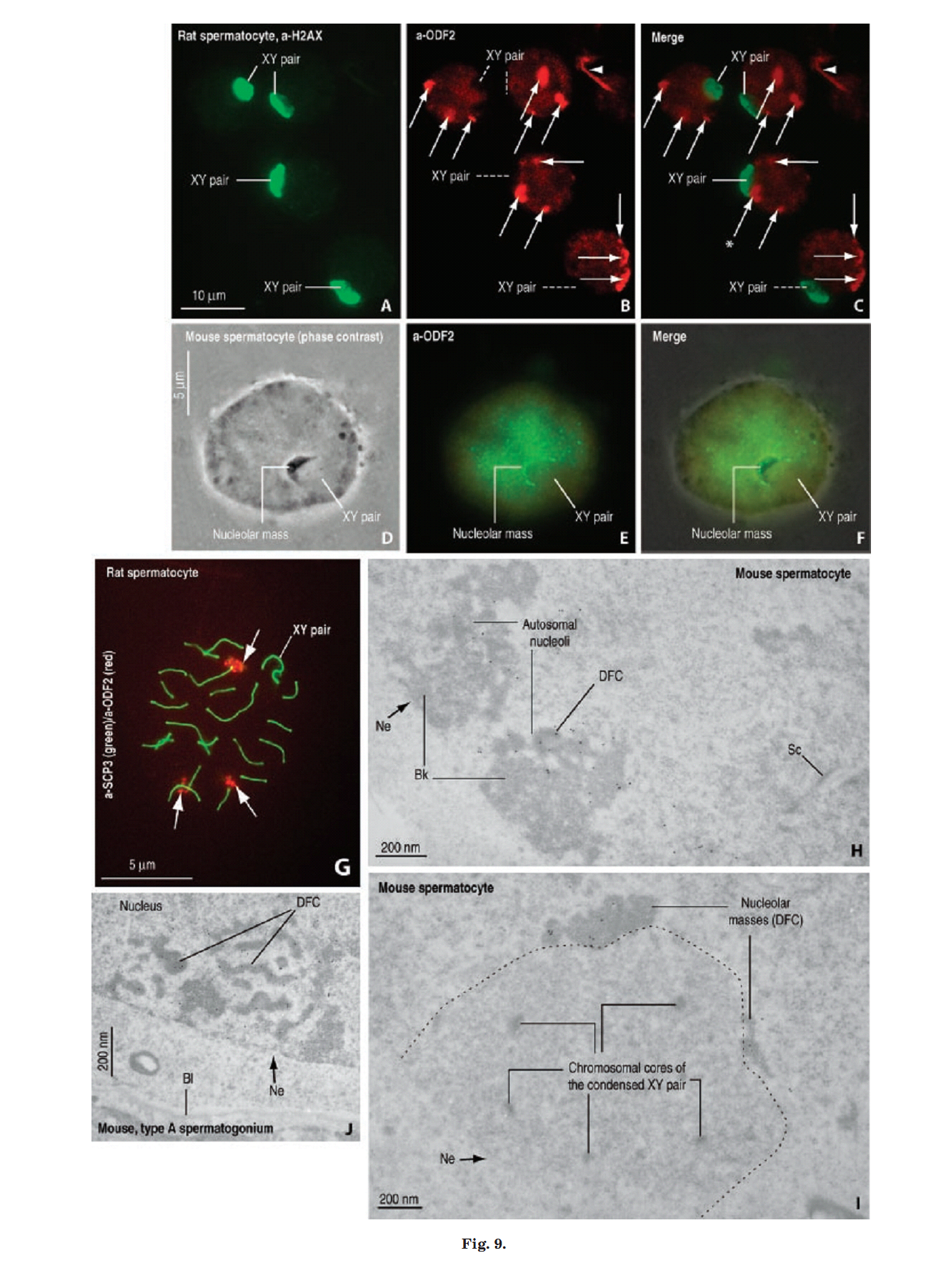
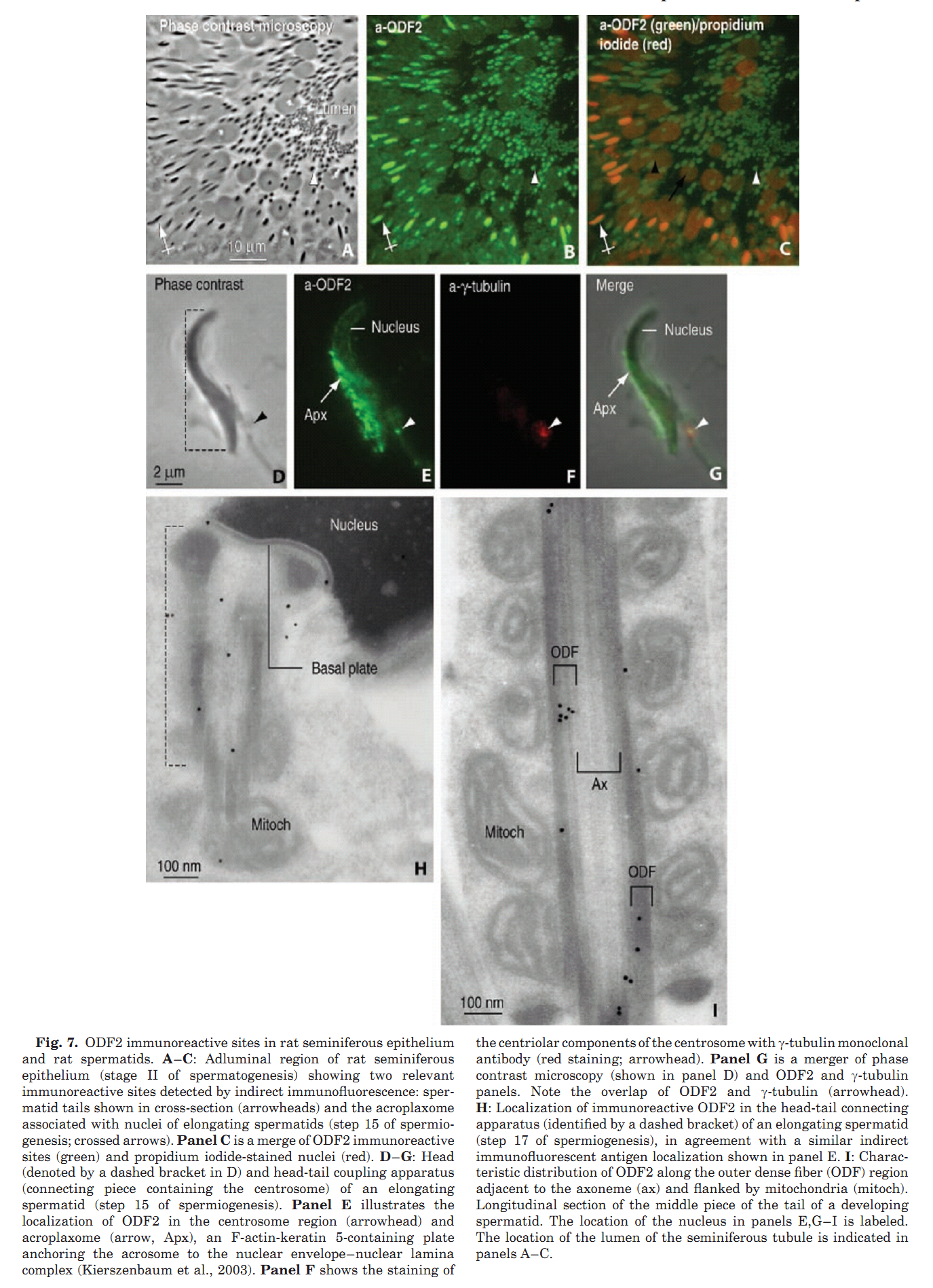
1. E. Rivkin, L. L. Tres and A. L. Kierszenbaum (2008) Genomic origin, processing and developmental expression of testicular outer dense fiber 2 (ODF2) transcripts and a novel nucleolar localization of ODF2 protein. Mol Reprod Dev 75(11): 1591-606. Abstract Outer dense fibers are a major constituent of the sperm tail and outer dense fiber 2 (ODF2) protein is one of their major components. ODF2 shares partial homology with cenexin 1 and cenexin 2, regarded as centriolar proteins. We show that ODF2 and cenexin 2 transcripts are the product of differential splicing of a single gene, designated Cenexin/ODF2 and that cenexin 1 is an incomplete clone of ODF2. ODF2 terminates in exon 20b whereas in cenexin 2 this exon is spliced out and translation terminates in exon 24. We demonstrate a transcriptional switch during rat testicular development, from somatic-type to testis-type ODF2 and cenexin transcripts during the onset of meiosis. The switch is completed when spermiogenesis is established. ODF2 immunoreactive sites were visualized in the acroplaxome, along the sperm tail and the centrosome-derived sperm head-to-tail coupling apparatus. An unexpected finding was the presence of ODF2 antigenic sites, but not cenexin antigenic sites, in the dense fibrillar component of the nucleolus of Sertoli cells, spermatogonia and primary spermatocytes. The characterization of the genomic origin, processing and developmental expression of ODF2 transcript isoforms and their protein products can help reconcile differences in the literature on the role of ODF2 and cenexin in the centrosome. Furthermore, the finding of ODF2 in the dense fibrillar component of the nucleolus suggests that this protein, in addition to its presence in sperm outer dense fibers and centrosome, highlights and adds to the nucleolar function during spermatogenesis and early embryogenesis. PMID: [18398819] Back to Top |
||||||||||||||||||||||||
Figures for illustrating the function of this protein/gene |
|
||||||||||||||||||||||||
Function |
Seems to be a major component of sperm tail outer densefibers (ODF). ODFs are filamentous structures located on theoutside of the axoneme in the midpiece and principal piece of themammalian sperm tail and may help to maintain the passive elasticstructures and elastic recoil of the sperm tail. May have amodulating influence on sperm motility. Functions as a generalscaffold protein that is specifically localized at thedistal/subdistal appendages of mother centrioles. Component of thecentrosome matrix required for the localization of PLK1 and NIN tothe centrosomes. Required for the formation and/or maintenance ofnormal CETN1 assembly. Back to Top |
||||||||||||||||||||||||
Subcellular Location |
Cytoplasm, cytoskeleton, centrosome. Cellprojection, cilium. Cytoplasm, cytoskeleton, centrosome,centriole. Cytoplasm, cytoskeleton, spindle pole. Note=Localizedat the microtubule organizing centers in interphase and spindlepoles in mitosis. Localized at the distal/subdistal appendages ofmother centrioles. |
||||||||||||||||||||||||
Tissue Specificity |
Testis-specific. Expressed in the proximalcompartment of the elongated spermatid tail; later expressionprogresses to the distal spermatid tail compartment located in thelumen of the seminiferous epithelium. In spermatids (stages II-III) expression of the tails peaks and remains strong during theremaining steps of spermiogenesis (at protein level). Expressioncorrelates with the onset of spermatogenesis and is first detectedat 30 days. Higher expression is seen in testis of 40-day-old andadults that are older than 50 days. No expression is seen in 10-and 20-day-old testes. |
||||||||||||||||||||||||
Gene Ontology |
|
||||||||||||||||||||||||
Interpro |
|||||||||||||||||||||||||
Pfam |
|||||||||||||||||||||||||
SMART |
|||||||||||||||||||||||||
PROSITE |
|||||||||||||||||||||||||
PRINTS |
|||||||||||||||||||||||||
Created Date |
18-Oct-2012 |
||||||||||||||||||||||||
Record Type |
Experiment identified |
||||||||||||||||||||||||
Protein sequence Annotation |
CHAIN 1 825 Outer dense fiber protein 2. /FTId=PRO_0000299460. COILED 139 212 Potential. COILED 240 418 Potential. COILED 456 793 Potential. VAR_SEQ 1 42 Missing (in isoform 4 and isoform 5). /FTId=VSP_027677. VAR_SEQ 1 36 MKDRSSTPPLHVHVDENTPVHVHIKKLPKPSAASSQ -> M SASSSGGSPRFPSCGKNGVTSLTQKKVLRTPCGAPSVTVT (in isoform 3). /FTId=VSP_027678. VAR_SEQ 60 78 Missing (in isoform 3, isoform 4 and isoform 5). /FTId=VSP_027679. VAR_SEQ 276 276 E -> ELCLKVPECARQHRPGRERQEDCQ (in isoform 5). /FTId=VSP_027680. VAR_SEQ 630 652 RSRIEHQGDKLELAREKHQASQK -> HSRVRDWQKGSHEL ARAGARLPR (in isoform 5). /FTId=VSP_027681. VAR_SEQ 633 652 IEHQGDKLELAREKHQASQK -> VRDWQKGSHELARAGAR LPR (in isoform 3 and isoform 4). /FTId=VSP_027682. VAR_SEQ 653 825 Missing (in isoform 3, isoform 4 and isoform 5). /FTId=VSP_027683. VAR_SEQ 763 769 VDRRYQS -> ENQKCWK (in isoform 2). /FTId=VSP_027684. VAR_SEQ 770 825 Missing (in isoform 2). /FTId=VSP_027685. CONFLICT 46 46 D -> Y (in Ref. 2; AAC53134). CONFLICT 107 108 DD -> EE (in Ref. 1; CAA64567). CONFLICT 120 120 K -> E (in Ref. 2; AAC53134). CONFLICT 483 483 D -> H (in Ref. 2; AAC53134). CONFLICT 511 511 A -> P (in Ref. 3; AAC08408). CONFLICT 543 543 S -> T (in Ref. 1; CAA64567). Back to Top |
||||||||||||||||||||||||
Nucleotide Sequence |
Length: 2203 bp Go to nucleotide: FASTA |
||||||||||||||||||||||||
Protein Sequence |
Length: 825 bp Go to amino acid: FASTA |
||||||||||||||||||||||||
The verified Protein-Protein interaction information |
| ||||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
||||||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
||||||||||||||||||||||||
Comments |
|||||||||||||||||||||||||