| Tag | Content | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00002000 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
UniProt Accession |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Theoretical PI |
6.13
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Weight |
52239 Da
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genbank Nucleotide ID |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genbank Protein ID |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Name |
Smad2 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Synonyms/Alias |
Madh2 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Name |
Mothers against decapentaplegic homolog 2 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Synonyms/Alias |
MAD homolog 2Mothers against DPP homolog 2 Mad-related protein 2; SMAD family member 2;SMAD 2Smad2 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Organism |
Rattus norvegicus (Rat) |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
NCBI Taxonomy ID |
10116 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Chromosome Location |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function in Stage |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function in Cell Type |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Description |
Temporarily unavailable |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The information of related literatures |
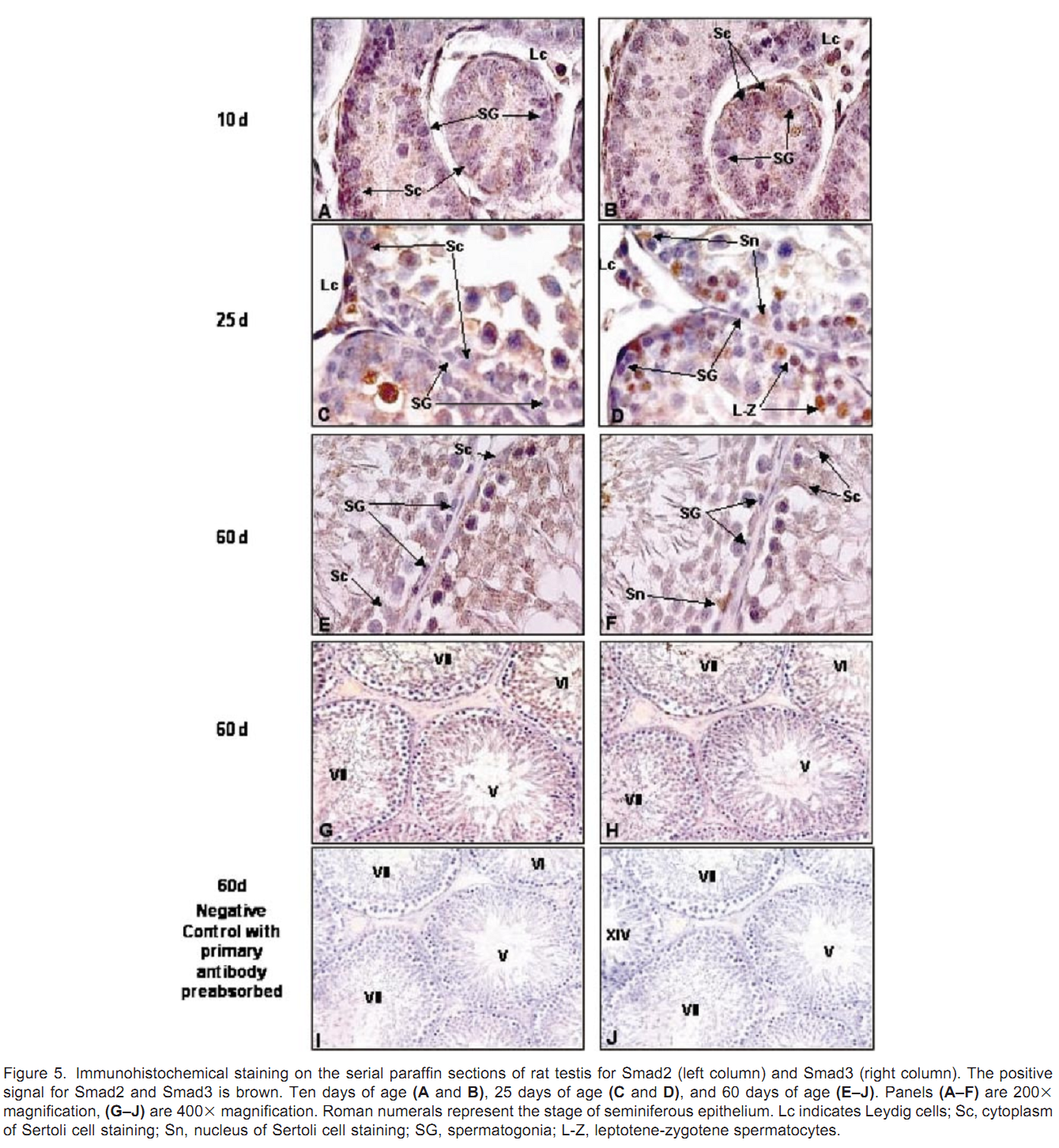
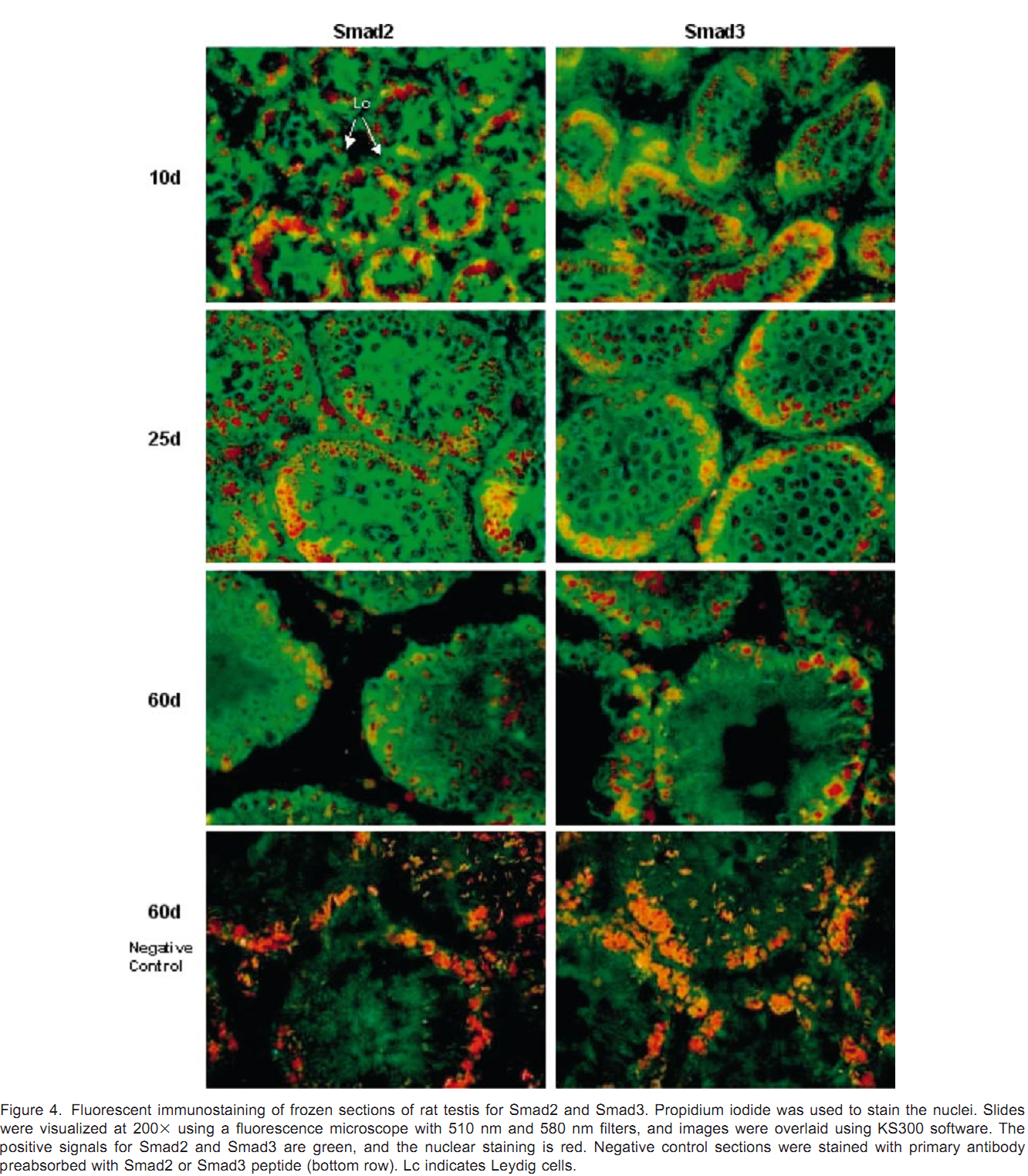
1. J. Xu, A. R. Beyer, W. H. Walker and E. A. McGee (2003) Developmental and stage-specific expression of Smad2 and Smad3 in rat testis. J Androl 24(2): 192-200. Abstract Members of the transforming growth factor beta type (TGFbeta) superfamily and their receptors are expressed in the testis, and are believed to play important paracrine and autocrine roles during testicular development and spermatogenesis. The Smad proteins are downstream mediators for the family of TGFbeta growth factors. Smad2 and Smad3 are associated with both TGFbeta and activin signaling. However, very little is known about the expression and regulation of the Smad signaling proteins in the testis. In the present study, we have determined that Smad2 and Smad3 proteins are expressed in the postnatal testes of rats from 5 days to 60 days of age. Expression levels for both proteins are higher in young rats than in sexually mature rats. Smad2 and Smad3 messenger RNA levels parallel protein expression. Smad2 and Smad3 proteins are mainly localized in the cytoplasm of meiotic germ cells, Sertoli cells, and Leydig cells. Smad3 protein is localized to the nucleus of preleptotene to zygotene primary spermatocytes in young rats. Both proteins are expressed throughout all stages of the cycle of seminiferous tubules but are expressed at their lowest levels at stages VII-VIII in the seminiferous epithelium of adult rats. The presence of these downstream mediators in these cell types supports a role for TGFbeta and activin during spermatogenesis. The difference between the expression of Smad2 and Smad3 suggests that they may have different functions within the testis. PMID: [12634305] Back to Top |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figures for illustrating the function of this protein/gene |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Function |
Receptor-regulated SMAD (R-SMAD) that is anintracellular signal transducer and transcriptional modulatoractivated by TGF-beta (transforming growth factor) and activintype 1 receptor kinases. Binds the TRE element in the promoterregion of many genes that are regulated by TGF-beta and, onformation of the SMAD2/SMAD4 complex, activates transcription. Mayact as a tumor suppressor in colorectal carcinoma. Positivelyregulates PDPK1 kinase activity by stimulating its dissociationfrom the 14-3-3 protein YWHAQ which acts as a negative regulator(By similarity). Back to Top |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subcellular Location |
Cytoplasm (By similarity). Nucleus (Bysimilarity). Note=Cytoplasmic and nuclear in the absence of TGF-beta. On TGF-beta stimulation, migrates to the nucleus whencomplexed with SMAD4. On dephosphorylation by phosphatase PPM1A,released from the SMAD2/SMAD4 complex, and exported out of thenucleus by interaction with RANBP1 (By similarity). |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tissue Specificity |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene Ontology |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Interpro |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Pfam |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
SMART |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PROSITE |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
PRINTS |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Created Date |
18-Oct-2012 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Record Type |
Experiment identified |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein sequence Annotation |
INIT_MET 1 1 Removed (By similarity). CHAIN 2 467 Mothers against decapentaplegic homolog 2. /FTId=PRO_0000090854. DOMAIN 10 176 MH1. DOMAIN 274 467 MH2. MOTIF 221 225 PY-motif (By similarity). MOD_RES 2 2 N-acetylserine (By similarity). MOD_RES 8 8 Phosphothreonine (By similarity). MOD_RES 19 19 N6-acetyllysine (By similarity). MOD_RES 240 240 Phosphoserine; by CAMK2 (By similarity). MOD_RES 458 458 Phosphoserine (By similarity). MOD_RES 464 464 Phosphoserine (By similarity). MOD_RES 465 465 Phosphoserine; by TGFBR1 (By similarity). MOD_RES 467 467 Phosphoserine; by TGFBR1 (By similarity). Back to Top |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Nucleotide Sequence |
Length: 1942 bp Go to nucleotide: FASTA |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Sequence |
Length: 467 bp Go to amino acid: FASTA |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The verified Protein-Protein interaction information |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Comments |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||