| Tag | Content | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
SG ID |
SG00002046 |
||||||||||||||||||||||||
UniProt Accession |
|||||||||||||||||||||||||
Theoretical PI |
5.14
|
||||||||||||||||||||||||
Molecular Weight |
93439 Da
|
||||||||||||||||||||||||
Genbank Nucleotide ID |
|||||||||||||||||||||||||
Genbank Protein ID |
|||||||||||||||||||||||||
Gene Name |
Pde4a |
||||||||||||||||||||||||
Gene Synonyms/Alias |
|||||||||||||||||||||||||
Protein Name |
cAMP-specific 3',5'-cyclic phosphodiesterase 4A |
||||||||||||||||||||||||
Protein Synonyms/Alias |
EC=3.1.4.17 DPDE2; |
||||||||||||||||||||||||
Organism |
Rattus norvegicus (Rat) |
||||||||||||||||||||||||
NCBI Taxonomy ID |
10116 |
||||||||||||||||||||||||
Chromosome Location |
|
||||||||||||||||||||||||
Function in Stage |
|||||||||||||||||||||||||
Function in Cell Type |
|||||||||||||||||||||||||
Description |
Temporarily unavailable |
||||||||||||||||||||||||
The information of related literatures |
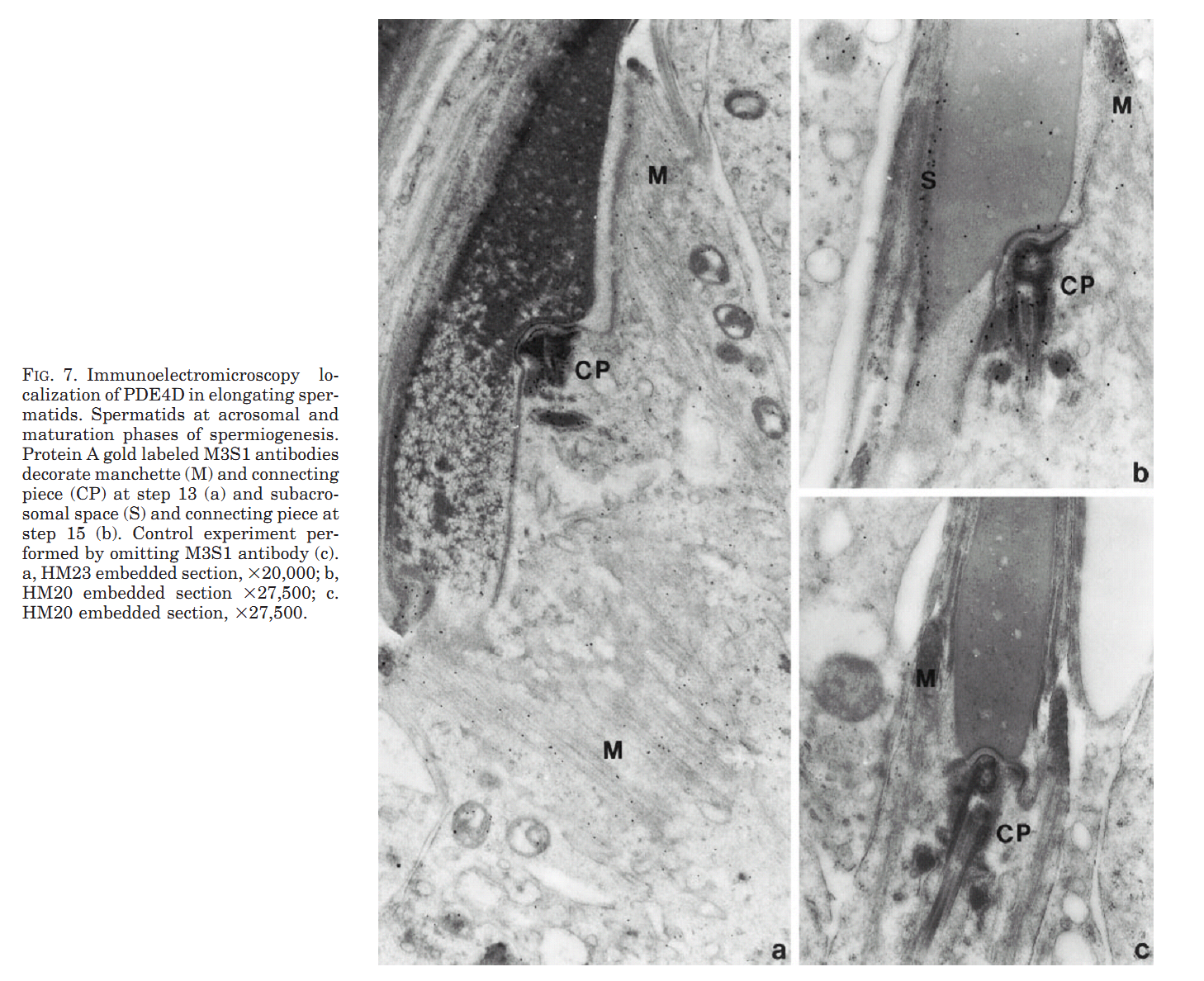
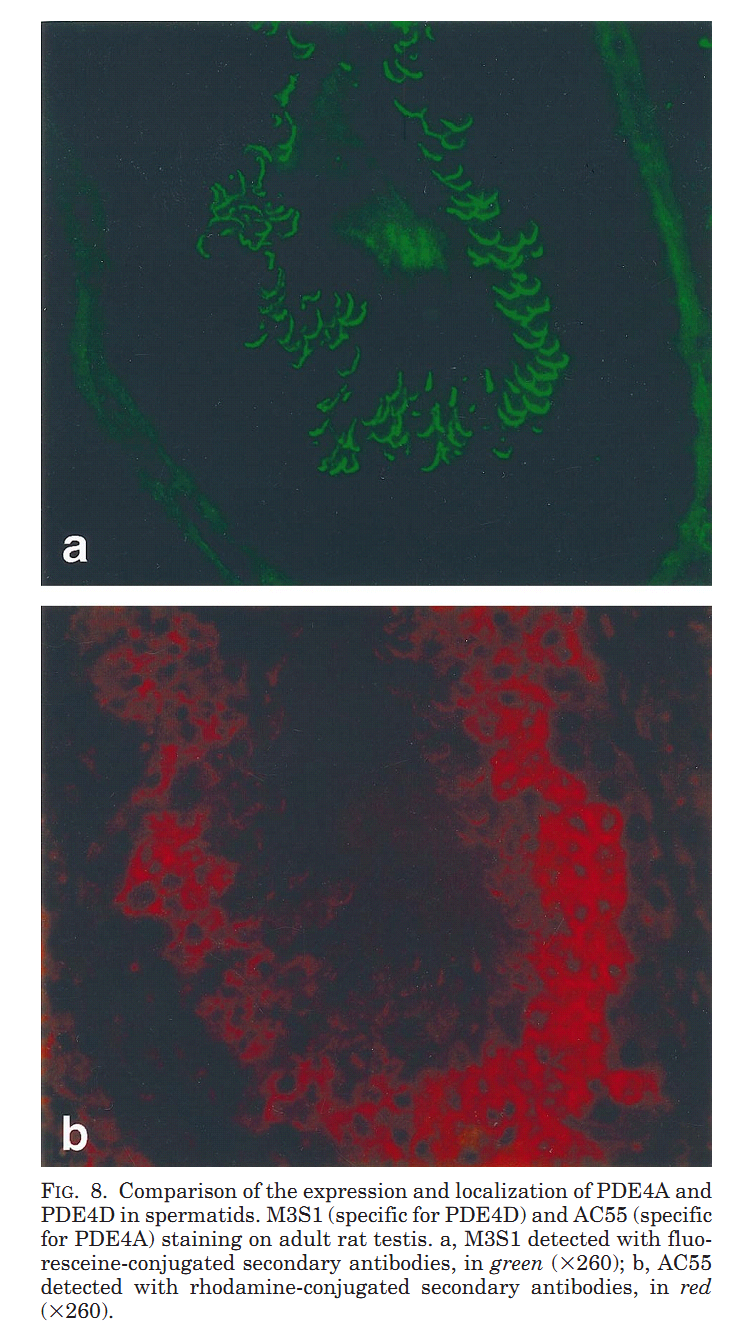
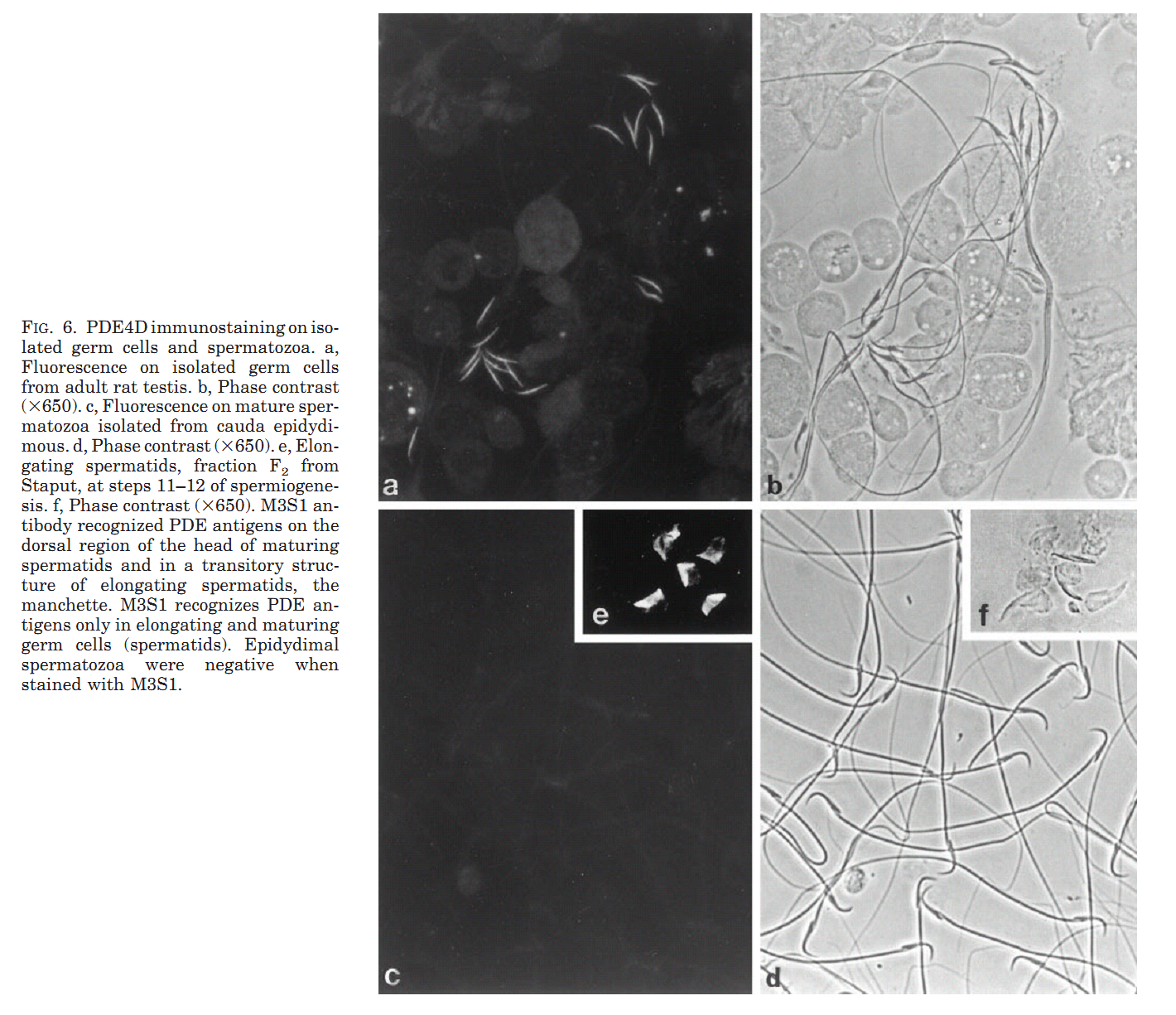
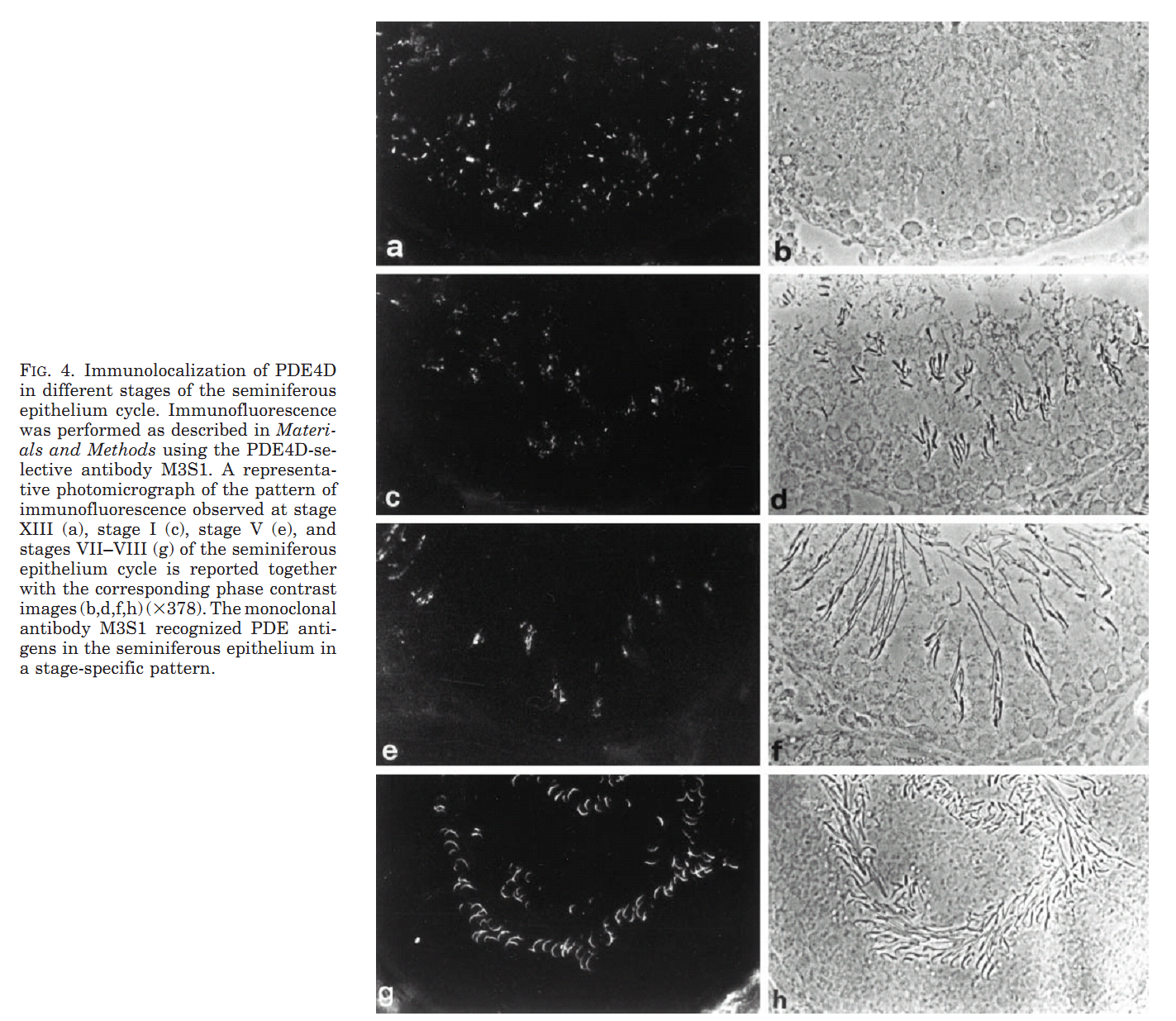
1. M. Salanova, S. Y. Chun, S. Iona, C. Puri, M. Stefanini and M. Conti (1999) Type 4 cyclic adenosine monophosphate-specific phosphodiesterases are expressed in discrete subcellular compartments during rat spermiogenesis. Endocrinology 140(5): 2297-306. Abstract The type 4 cAMP-specific phosphodiesterases (PDE4) are a family of closely related enzymes with similar catalytic domains and divergent amino- and carboxyl-terminus domains. Multiple PDE proteins with heterogeneous amino termini are derived from each gene. To understand the significance of this heterogeneity, the expression and localization of variants derived from PDE4A and PDE4D genes was investigated during spermatogenesis in the rat. RNase protection analysis with mRNA for testes at different ages of development showed that two transcripts (PDE4D1 and PDE4D2) are expressed at day 10 and 15 of age and become undetectable thereafter. An additional PDE4D transcript appears at day 30 and increased during testid maturation. This latter transcript codes for a long variant of the PDE4D gene and is expressed in germ cells as demonstrated by RNase protection with RNA from isolated pachytene spermatocytes and round spermatids. The presence of a corresponding PDE4D protein with a molecular mass of 98 kDa was established by immunoprecipitation and Western blot analysis with antibodies specific for PDE4D and by immunoaffinity chromatography purification of the 98 kDa variant from isolated germ cells. PDE4A transcripts were also expressed in pachytene spermatocytes and round spermatids. Two polypeptides encoded by these PDE4A transcripts were expressed in pachytene spermatocytes, reached a maximum in round spermatids, and declined thereafter. Immunofluorescence analysis demonstrated a localization of the PDE4D protein in the manchette and in a periacrosomal region of the developing spermatid, a localization confirmed by immunogold electron microscopy. Conversely, the PDE4A was mostly soluble in the cytoplasm of round spermatids. These data demonstrate that PDE4D and PDE4A variants are expressed at different stages and localized in distinct subcellular structures of developing spermatids. Different properties of the mRNAs derived from the two genes and localization signals are responsible for the temporal and spatial expression of the different PDE4 isoenzymes. PMID: [10218983] Back to Top |
||||||||||||||||||||||||
Figures for illustrating the function of this protein/gene |
|
||||||||||||||||||||||||
Function |
Hydrolyzes the second messenger cAMP, which is a keyregulator of many important physiological processes (Bysimilarity). Back to Top |
||||||||||||||||||||||||
Subcellular Location |
Cytoplasm, perinuclear region (Bysimilarity). |
||||||||||||||||||||||||
Tissue Specificity |
Isoform 2 is testis specific. |
||||||||||||||||||||||||
Gene Ontology |
|
||||||||||||||||||||||||
Interpro |
IPR003607; HD/PDEase_dom. IPR023088; PDEase. IPR002073; PDEase_catalytic_dom. IPR023174; PDEase_CS. Back to Top |
||||||||||||||||||||||||
Pfam |
|||||||||||||||||||||||||
SMART |
|||||||||||||||||||||||||
PROSITE |
|||||||||||||||||||||||||
PRINTS |
|||||||||||||||||||||||||
Created Date |
18-Oct-2012 |
||||||||||||||||||||||||
Record Type |
Experiment identified |
||||||||||||||||||||||||
Protein sequence Annotation |
CHAIN 1 844 cAMP-specific 3',5'-cyclic phosphodiesterase 4A. /FTId=PRO_0000198808. NP_BIND 419 423 cAMP (By similarity). REGION 317 710 Catalytic (By similarity). ACT_SITE 419 419 Proton donor (By similarity). METAL 423 423 Divalent metal cation 1 (By similarity). METAL 459 459 Divalent metal cation 1 (By similarity). METAL 460 460 Divalent metal cation 1 (By similarity). METAL 460 460 Divalent metal cation 2 (By similarity). METAL 577 577 Divalent metal cation 1 (By similarity). BINDING 460 460 cAMP (By similarity). BINDING 577 577 cAMP (By similarity). BINDING 628 628 cAMP (By similarity). SITE 69 70 Cleavage; by caspase-3 (By similarity). SITE 580 580 Binds AMP, but not cAMP (By similarity). MOD_RES 147 147 Phosphoserine; by MAPKAPK2. MOD_RES 160 160 Phosphoserine (By similarity). MOD_RES 672 672 Phosphoserine (By similarity). MOD_RES 674 674 Phosphoserine (By similarity). CROSSLNK 344 344 Glycyl lysine isopeptide (Lys-Gly) (interchain with G-Cter in SUMO) (By similarity). VAR_SEQ 1 318 Missing (in isoform 5). /FTId=VSP_004569. VAR_SEQ 1 259 Missing (in isoform 4). /FTId=VSP_004568. VAR_SEQ 1 234 Missing (in isoform 3). /FTId=VSP_004566. VAR_SEQ 1 102 MEPPAAPSERSLSLSLPGPREGQATLKPPPQHLWRQPRTPI RIQQRGYPDSAERSETERSPHRPIERADAVDTGDRPGLRTT RMSWPSSFHGTGTGGGSSRR -> MPSRKRLTLPRIFIVRK NGNS (in isoform 2). /FTId=VSP_004565. VAR_SEQ 1 102 MEPPAAPSERSLSLSLPGPREGQATLKPPPQHLWRQPRTPI RIQQRGYPDSAERSETERSPHRPIERADAVDTGDRPGLRTT RMSWPSSFHGTGTGGGSSRR -> ALPLGPESLTHFSFSEE DTLRHPPGRCVS (in isoform 6). /FTId=VSP_038187. VAR_SEQ 235 256 WCLEQLETMQTYRSVSEMASHK -> MPLVDFFCETCSKPW LVGWWDQ (in isoform 3). /FTId=VSP_004567. VAR_SEQ 354 386 Missing (in isoform 5). /FTId=VSP_004570. MUTAGEN 147 147 S->A: Abolishes phosphorylation by MAPKAPK2. MUTAGEN 161 161 S->A: Does not affect phosphorylation by MAPKAPK2. CONFLICT 130 130 A -> R (in Ref. 5; AAF14352). CONFLICT 465 466 GV -> AL (in Ref. 4; AAA41848/AAA41823). CONFLICT 603 604 GD -> AH (in Ref. 4; AAA41848/AAA41823). CONFLICT 833 833 A -> T (in Ref. 1; AAC37699/AAA41101/ AAA41102 and 5; AAF14352). HELIX 781 787 Back to Top |
||||||||||||||||||||||||
Nucleotide Sequence |
Length: 2914 bp Go to nucleotide: FASTA |
||||||||||||||||||||||||
Protein Sequence |
Length: 844 bp Go to amino acid: FASTA |
||||||||||||||||||||||||
The verified Protein-Protein interaction information |
| ||||||||||||||||||||||||
Other Protein-Protein interaction resources |
String database |
||||||||||||||||||||||||
View Microarray data |
Temporarily unavailable |
||||||||||||||||||||||||
Comments |
|||||||||||||||||||||||||