 Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
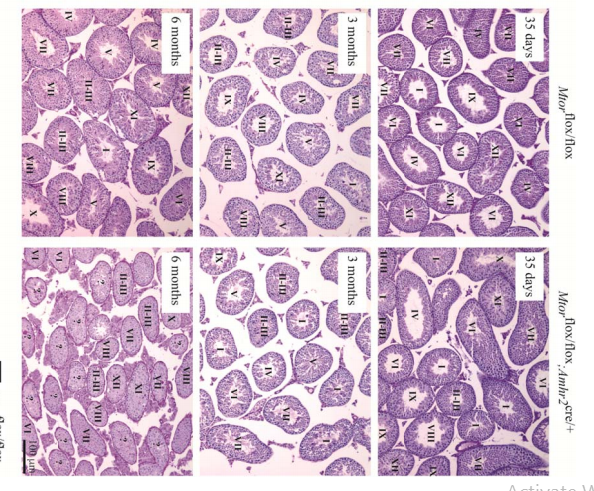
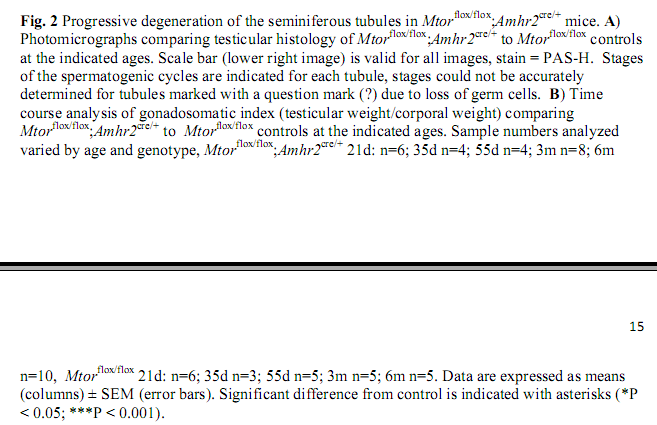 Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
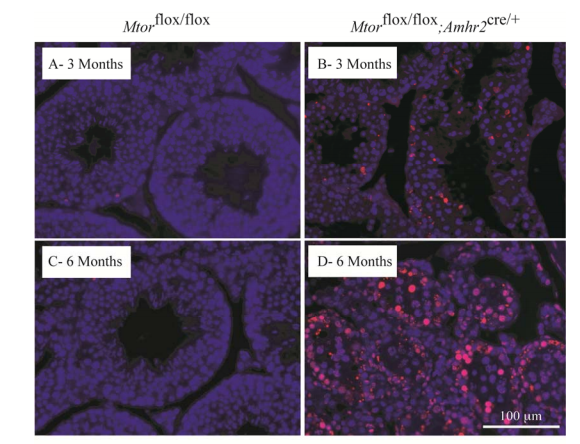 Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
 Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
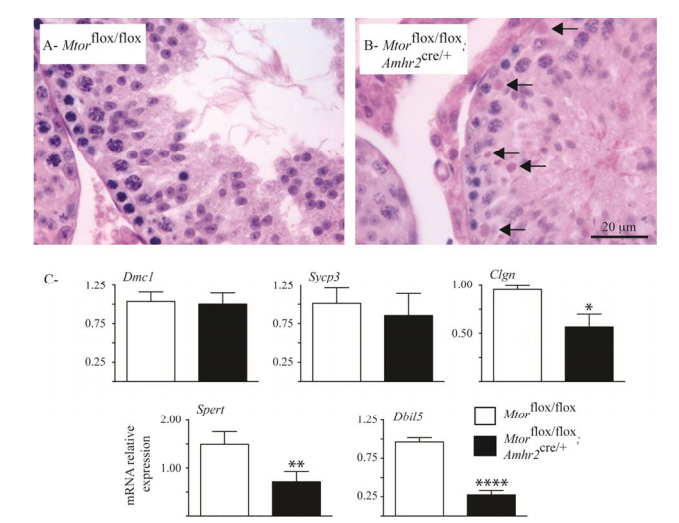 Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
 Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
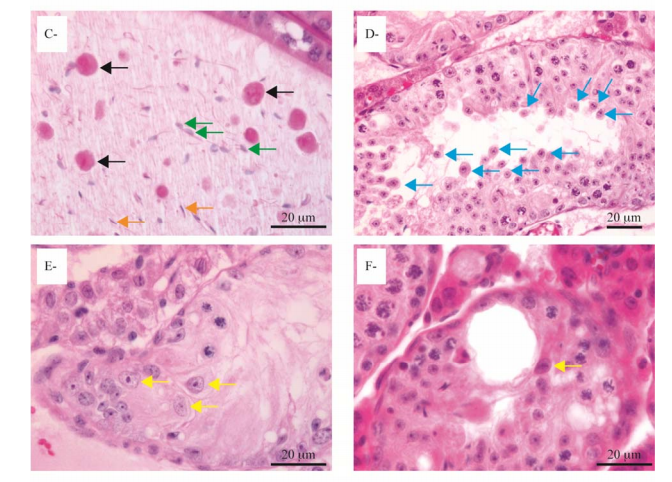 Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
 Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
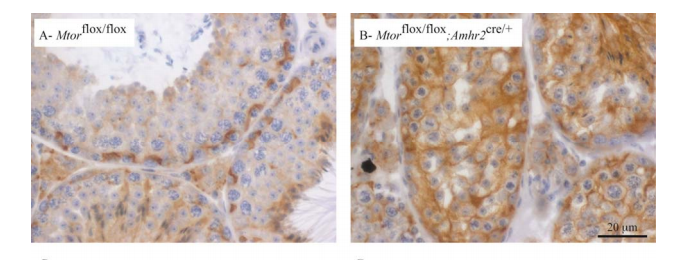 Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
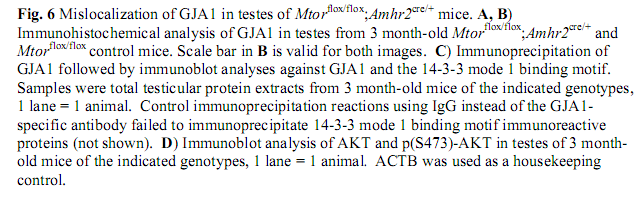 Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]
Boyer, A., Girard, M., Thimmanahalli, D. S., Levasseur, A., Celeste, C., Paquet, M., Duggavathi, R. and Boerboom, D. (2016) mTOR Regulates Gap Junction Alpha-1 Protein Trafficking in Sertoli Cells and Is Required for the Maintenance of Spermatogenesis in Mice. Biol Reprod 95(1): 13. PMID: [27281705]