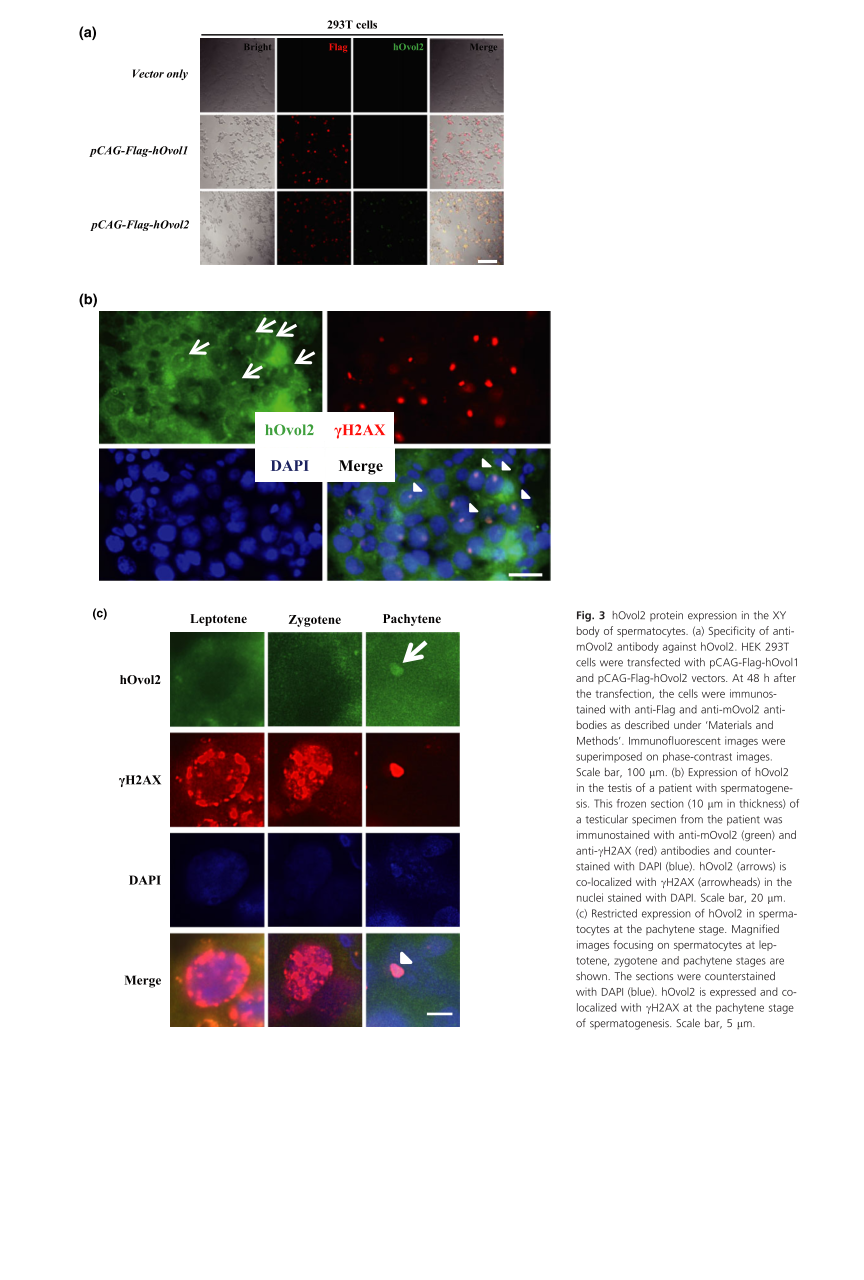
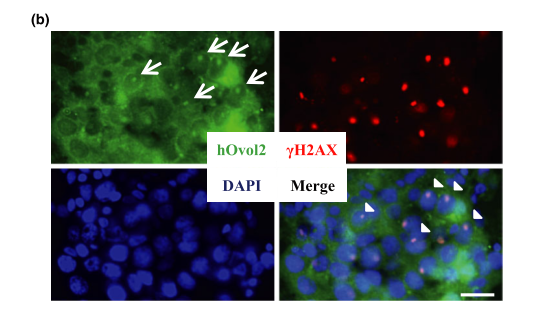
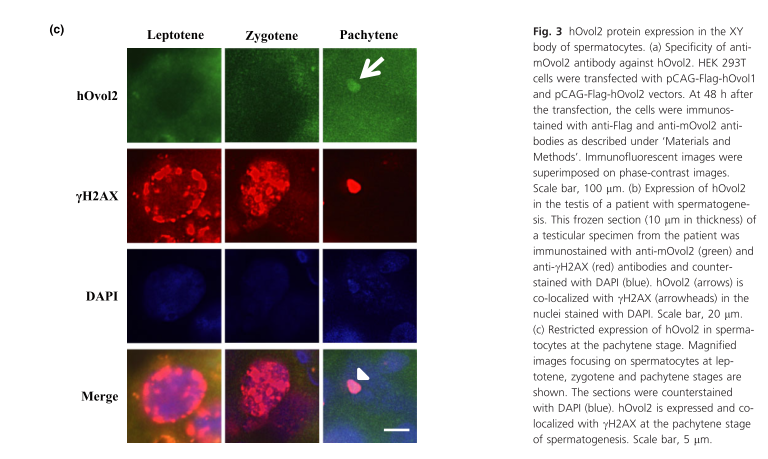
General information
| SG000008 | |||||||||||||||||
| OVOL2 | |||||||||||||||||
| Homo sapiens | |||||||||||||||||
| 9606 | |||||||||||||||||
| ENSG00000125850 | |||||||||||||||||
| ENSP00000278780 | |||||||||||||||||
Ovo like zinc finger 2 [Homo sapiens(human) ] | |||||||||||||||||
| |||||||||||||||||
Reviewed functional gene |
Functional information
| premeiotic meiotic | |
| Spermatocyte | |
|
1. Abstract | |
| |
| HOvol2 gene expressed in the testis, specifically in the XY body of spermatocytes at the pachytene stage. | |
| Corneal dystrophy posterior polymorphous 1 | |
| 616441 | |
| SCO |
Expression and location
| Expressed highest in stomach | |
| Expressed highest in spermatocyte | |
| View detail | |
| View detail | |
| Nucleus |
Mutation
|
1000G (Phase 3): 1132 ESP6500 (SI-V2): 49 ExAC (r0.3.1): 211 dbSNP(Build 147): 2206 |
|
|
Chinese health control (254): 4 European health control (283): 1 Chinese patients (168): |
|
| Gene Name: OVOL2 DNMR-average: 0.0000148075 DNMR-SC: 0.0000119297 |
Other information
|
Click the right sign for more information
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Q9BRP0 |