General information
| SG000020 | |||||||||||||||||
| NPAS2 | |||||||||||||||||
| MOP4, PASD4, bHLHe9 | |||||||||||||||||
| Homo sapiens | |||||||||||||||||
| 9606 | |||||||||||||||||
| ENSG00000170485 | |||||||||||||||||
| ENSP00000338283 ENSP00000397595 ENSP00000388528 ENSP00000395265 ENSP00000392125 ENSP00000393396 | |||||||||||||||||
Neuronal PAS domain protein 2 [Homo sapiens (human)] | |||||||||||||||||
| |||||||||||||||||
Reviewed functional gene |
Functional information
| meiotic | |
| Spermatid | |
|
1. Abstract | |
| |
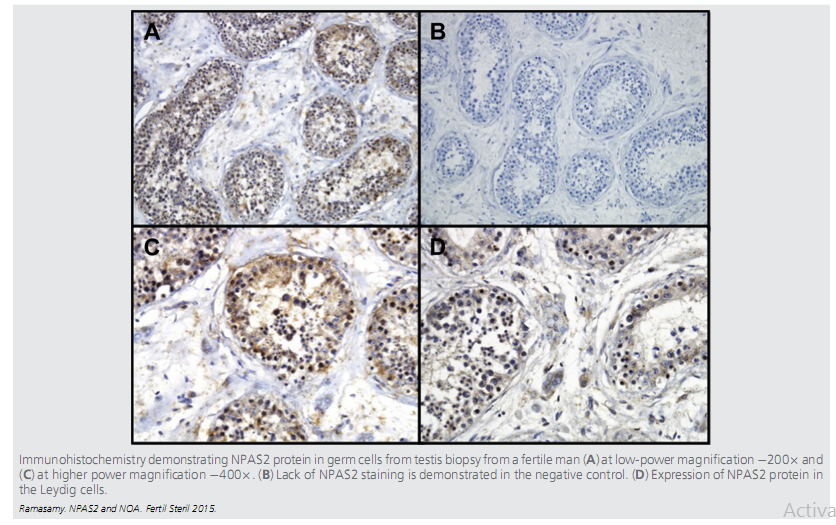
| Neuronal PAS 2 domain (NPAS2) protein is expressed in the brain, more specifically the cerebral cortex, testis, and the prostate in both mouse and human. This expression data is consistent with the role of NPAS2 function in circadian rhythm, fertility and suggest NPAS2 as one of the potential causative genes for NOA | |
|
ReactomeID: R-HSA-400253 Circadian Clock | |
| 603347 | |
| N/A |
Expression and location
| Expressed highest in esophagus | |
| Expressed highest in spermatogonium | |
| View detail | |
| View detail | |
| Nucleus |
Mutation
|
1000G (Phase 3): 4535 ESP6500 (SI-V2): 188 ExAC (r0.3.1): 831 dbSNP(Build 147): 8962 |
|
|
Chinese health control (254): 13 European health control (283): 5 Chinese patients (168): |
|
| Gene Name: NPAS2 DNMR-average: 0.0000402625 DNMR-SC: 0.0000419342 |
Other information
|
Click the right sign for more information
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Q99743 |