General information
| SG000028 | |||||||||||||||||
| KHDRBS1 | |||||||||||||||||
| Sam68, p62, p68 | |||||||||||||||||
| Homo sapiens | |||||||||||||||||
| 9606 | |||||||||||||||||
| ENSG00000121774 | |||||||||||||||||
| ENSP00000313829 ENSP00000417731 | |||||||||||||||||
KH RNA binding domain containing, signal transduction associated 1 [Homo sapiens (human)] | |||||||||||||||||
| |||||||||||||||||
Reviewed functional gene |
Functional information
| meiotic | |||||||||||||
| Spermatocyte | |||||||||||||
|
1. Abstract | |||||||||||||
| |||||||||||||
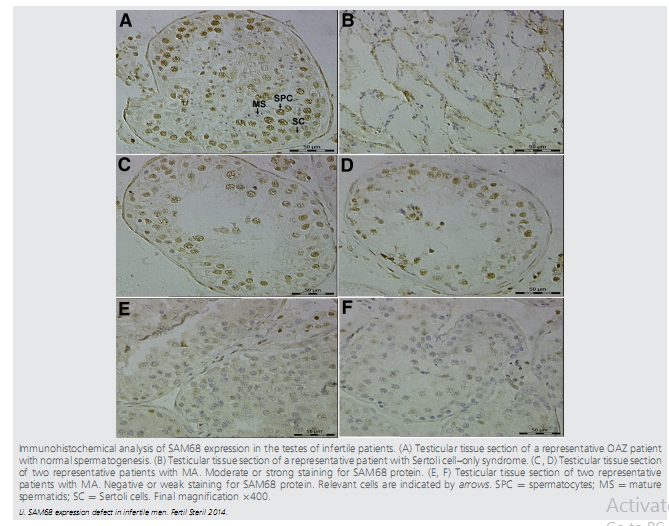
| Sam68 (Src-associated substrate in mitosis of 68 kD, also known as KH-DRBS1) belongs to the STAR (signal transduction and activation of RNA) family of RNA-binding proteins, which regulate a range of processes, including RNA stability, export, splicing, and mRNA translation. Reproductive phenotype of Sam68 knockout mice revealed that males are completely infertile. Deficiency of SAM68 expression in the human testis is associated with MA at the spermatocyte stage and SCOS. | |||||||||||||
| |||||||||||||
|
ReactomeID: R-HSA-8849468 PTK6 Regulates Proteins Involved in RNA Processing | |||||||||||||
| 602489 | |||||||||||||
| SCO |
Expression and location
| Expressed highest in ovary | |
| Expressed highest in spermatogonium | |
| View detail | |
| View detail | |
| Nucleus |
Mutation
|
1000G (Phase 3): 835 ESP6500 (SI-V2): 58 ExAC (r0.3.1): 313 dbSNP(Build 147): 1907 |
|
|
Chinese health control (254): European health control (283): Chinese patients (168): |
|
| Gene Name: KHDRBS1 DNMR-average: 0.00002052 DNMR-SC: 0.0000180509 |
Other information
|
Click the right sign for more information
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Q07666 |