 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
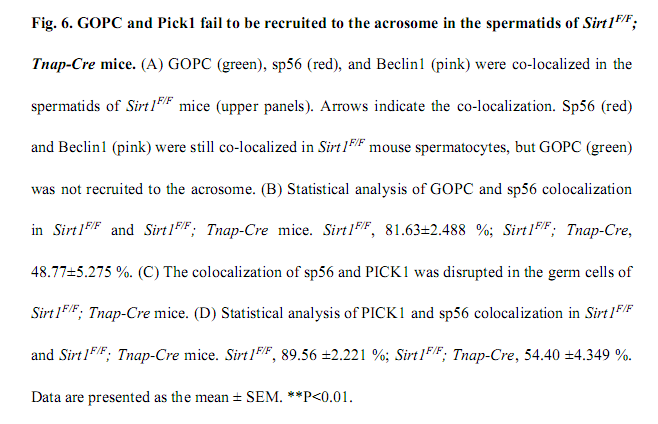 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
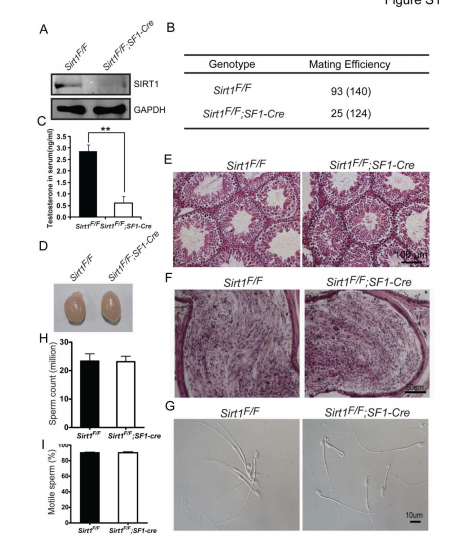 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
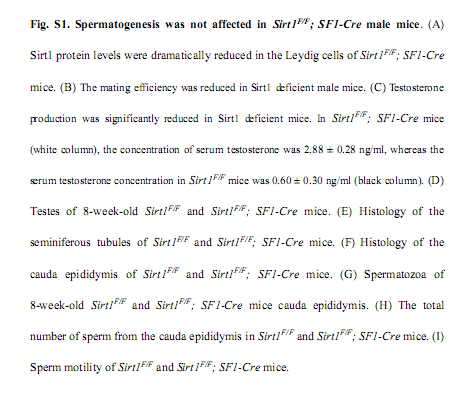 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
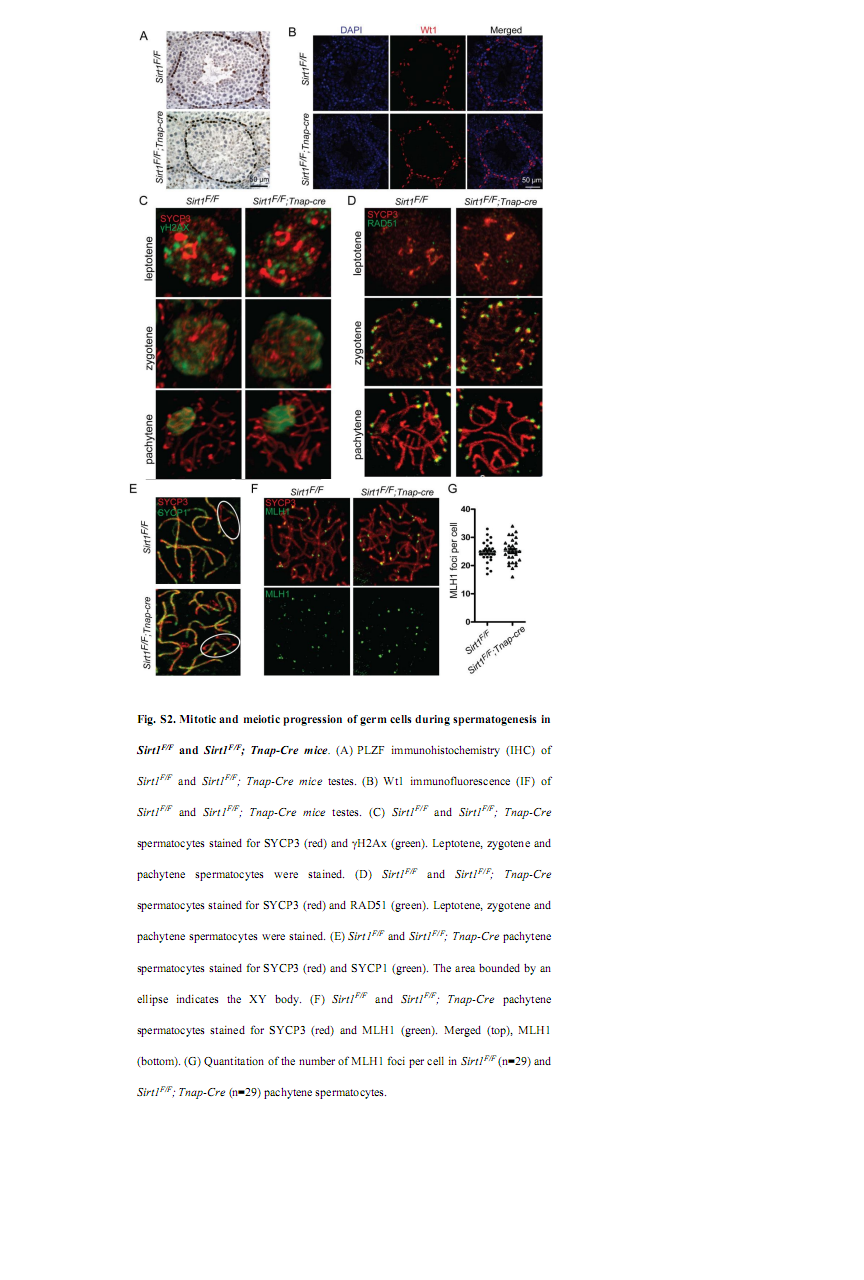 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
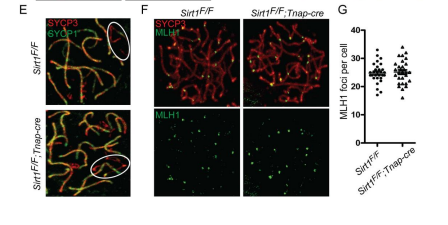 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
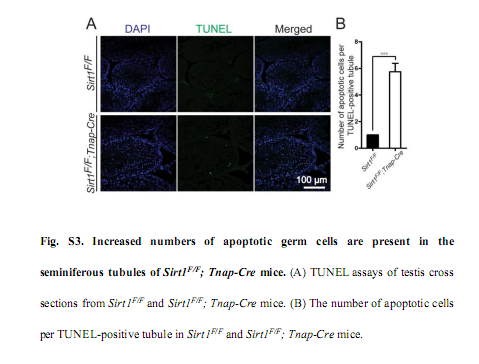 Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
Liu, C., Song, Z., Wang, L., Yu, H., Liu, W., Shang, Y., Xu, Z., Zhao, H., Gao, F., Wen, J., Zhao, L., Gui, Y., Jiao, J. and Li, W. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144(3): 441-451. PMID: [28003215]
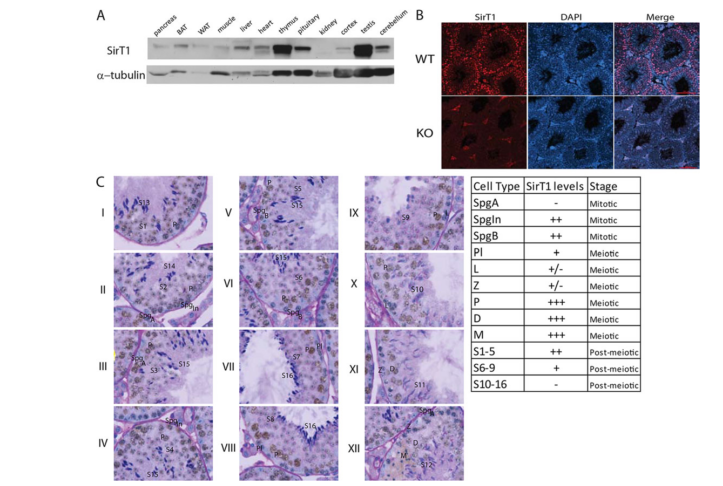 Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
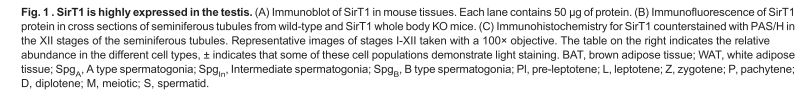 Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
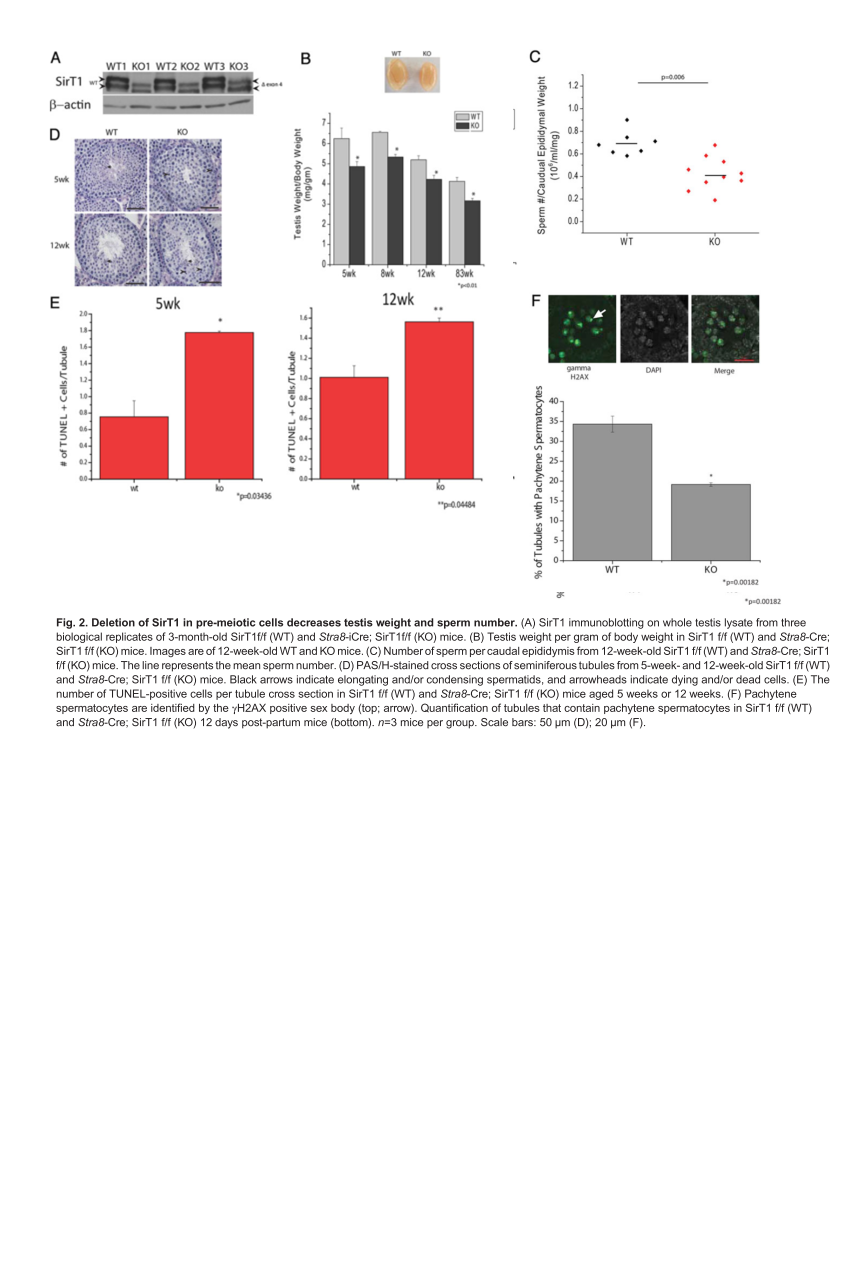 Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
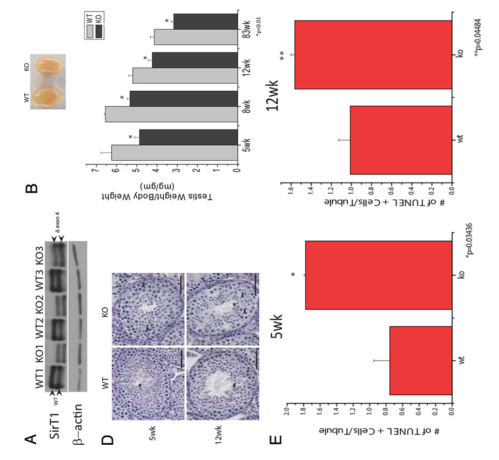 Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
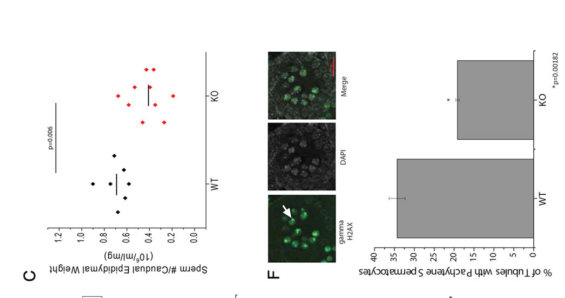 Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
 Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
 Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
 Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
 Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
 Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
 Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
 Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
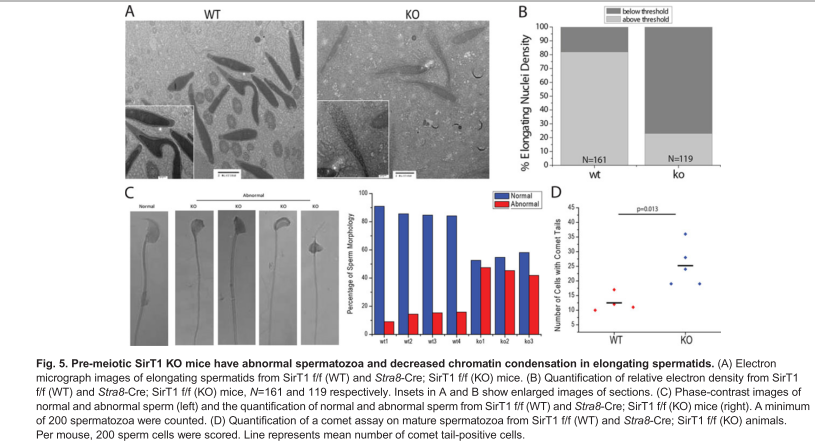 Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
Bell, E. L., Nagamori, I., Williams, E. O., Del Rosario, A. M., Bryson, B. D., Watson, N., White, F. M., Sassone-Corsi, P. and Guarente, L. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141(18): 3495-504. PMID: [25142464]
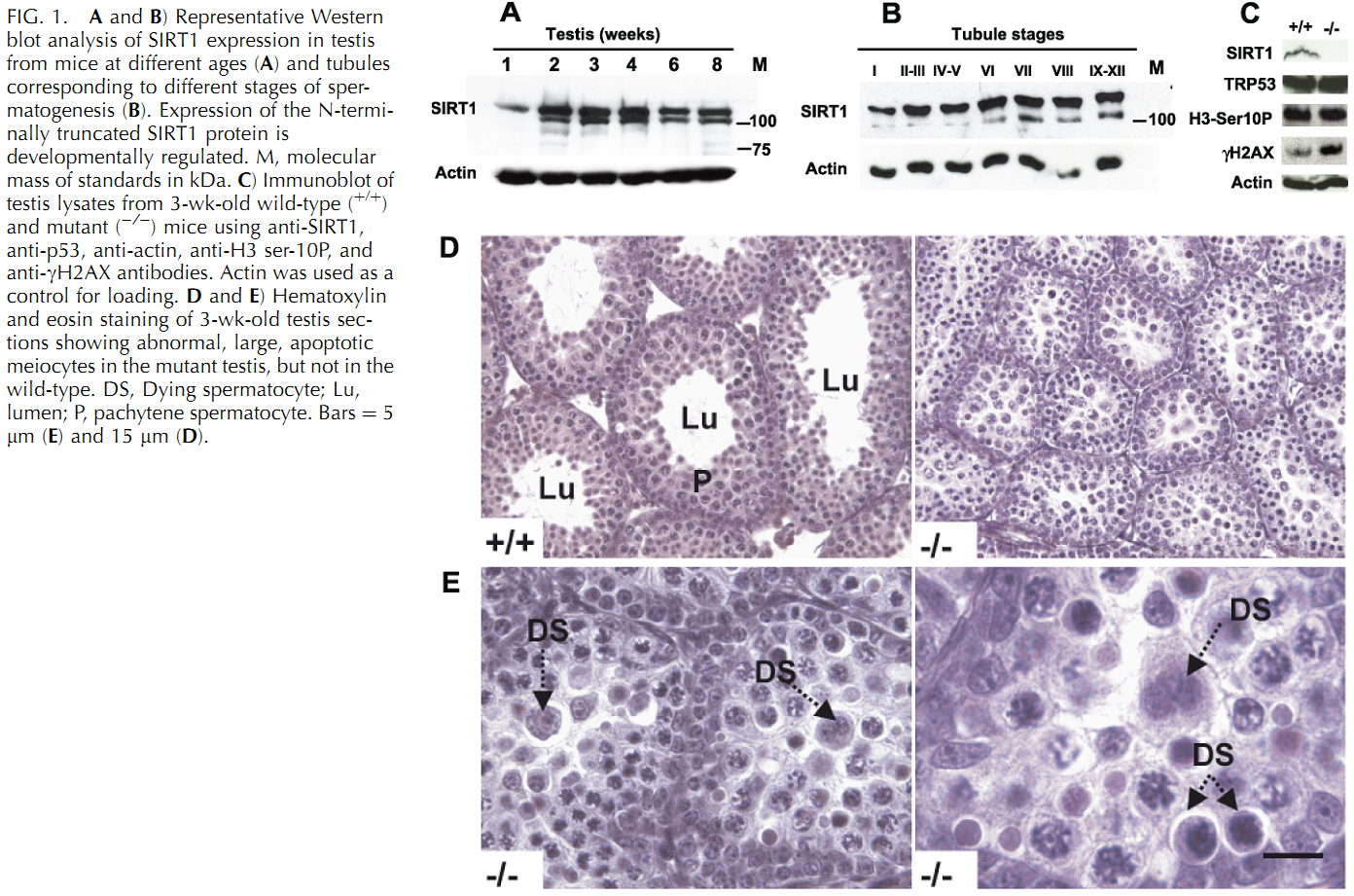 U. Kolthur-Seetharam, K. Teerds, D. G. de Rooij, O. Wendling, M. McBurney, P. Sassone-Corsi and I. Davidson (2009) The histone deacetylase SIRT1 controls male fertility in mice through regulation of hypothalamic-pituitary gonadotropin signaling. Biol Reprod 80(2): 384-91. PMID: [18987333]
U. Kolthur-Seetharam, K. Teerds, D. G. de Rooij, O. Wendling, M. McBurney, P. Sassone-Corsi and I. Davidson (2009) The histone deacetylase SIRT1 controls male fertility in mice through regulation of hypothalamic-pituitary gonadotropin signaling. Biol Reprod 80(2): 384-91. PMID: [18987333]
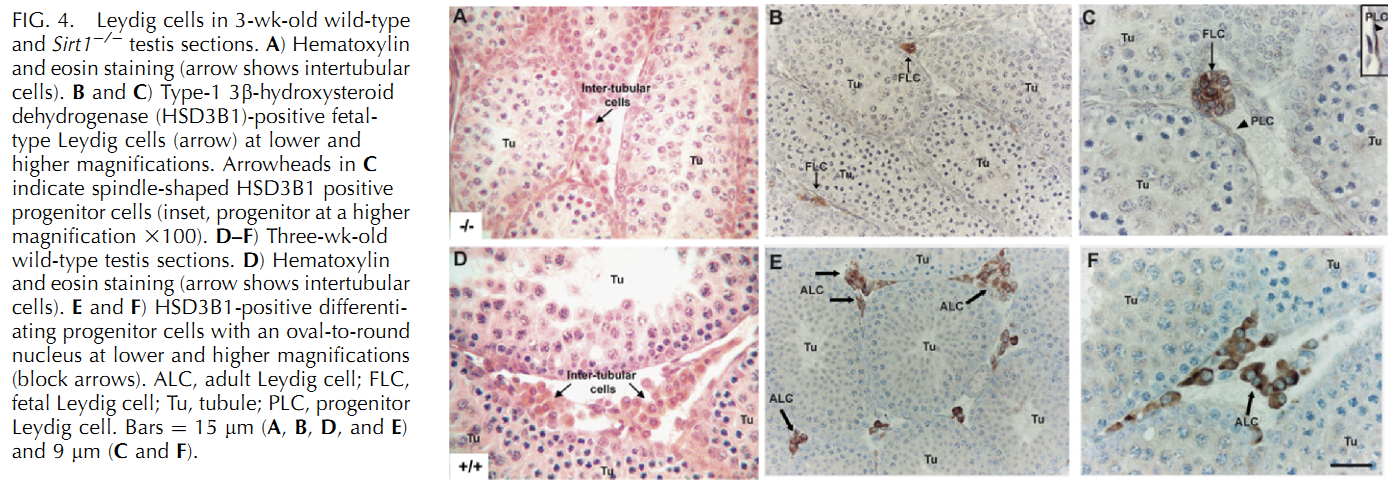 U. Kolthur-Seetharam, K. Teerds, D. G. de Rooij, O. Wendling, M. McBurney, P. Sassone-Corsi and I. Davidson (2009) The histone deacetylase SIRT1 controls male fertility in mice through regulation of hypothalamic-pituitary gonadotropin signaling. Biol Reprod 80(2): 384-91. PMID: [18987333]
U. Kolthur-Seetharam, K. Teerds, D. G. de Rooij, O. Wendling, M. McBurney, P. Sassone-Corsi and I. Davidson (2009) The histone deacetylase SIRT1 controls male fertility in mice through regulation of hypothalamic-pituitary gonadotropin signaling. Biol Reprod 80(2): 384-91. PMID: [18987333]
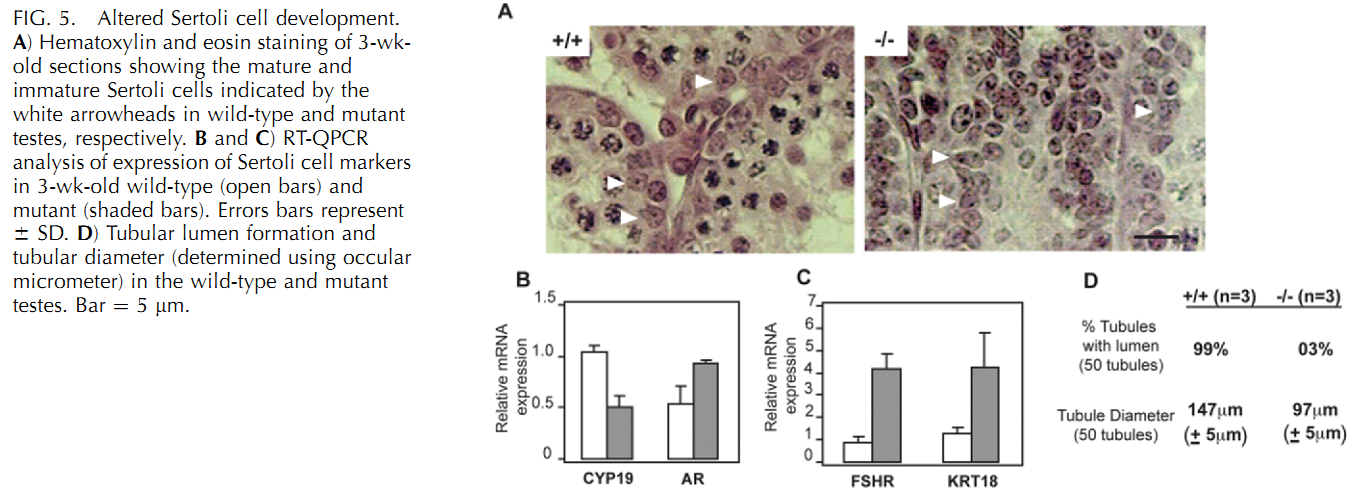 U. Kolthur-Seetharam, K. Teerds, D. G. de Rooij, O. Wendling, M. McBurney, P. Sassone-Corsi and I. Davidson (2009) The histone deacetylase SIRT1 controls male fertility in mice through regulation of hypothalamic-pituitary gonadotropin signaling. Biol Reprod 80(2): 384-91. PMID: [18987333]
U. Kolthur-Seetharam, K. Teerds, D. G. de Rooij, O. Wendling, M. McBurney, P. Sassone-Corsi and I. Davidson (2009) The histone deacetylase SIRT1 controls male fertility in mice through regulation of hypothalamic-pituitary gonadotropin signaling. Biol Reprod 80(2): 384-91. PMID: [18987333]
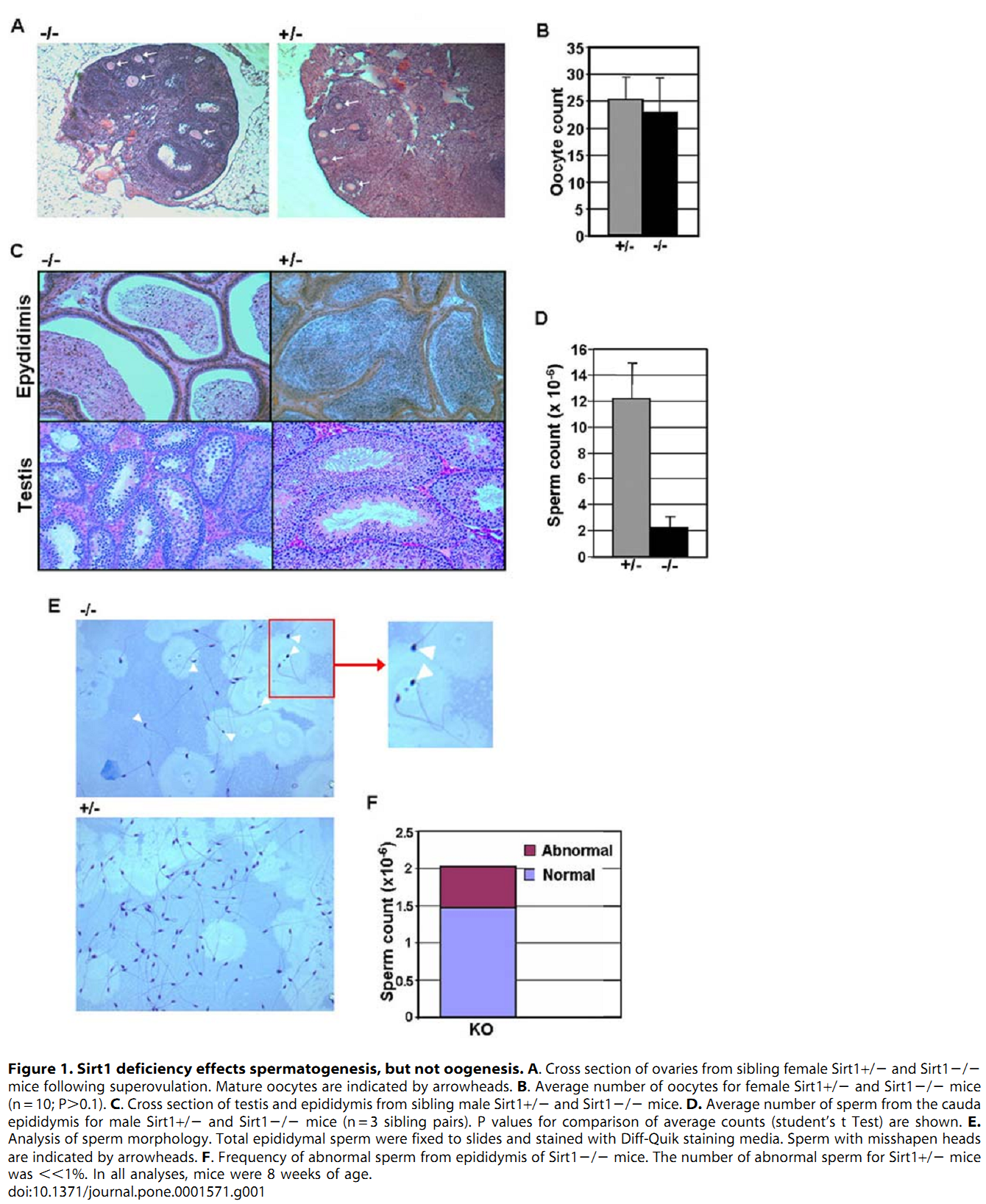 M. Coussens, J. G. Maresh, R. Yanagimachi, G. Maeda and R. Allsopp (2008) Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS One 3(2): e1571. PMID: [18270565]
M. Coussens, J. G. Maresh, R. Yanagimachi, G. Maeda and R. Allsopp (2008) Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS One 3(2): e1571. PMID: [18270565]





































