 Schrade, A., Kyronlahti, A., Akinrinade, O., Pihlajoki, M., Fischer, S., Rodriguez, V. M., Otte, K., Velagapudi, V., Toppari, J., Wilson, D. B. and Heikinheimo, M. (2016) GATA4 Regulates Blood-Testis Barrier Function and Lactate Metabolism in Mouse Sertoli Cells. Endocrinology 157(6): 2416-31. PMID: [26974005]
Schrade, A., Kyronlahti, A., Akinrinade, O., Pihlajoki, M., Fischer, S., Rodriguez, V. M., Otte, K., Velagapudi, V., Toppari, J., Wilson, D. B. and Heikinheimo, M. (2016) GATA4 Regulates Blood-Testis Barrier Function and Lactate Metabolism in Mouse Sertoli Cells. Endocrinology 157(6): 2416-31. PMID: [26974005]
 Schrade, A., Kyronlahti, A., Akinrinade, O., Pihlajoki, M., Fischer, S., Rodriguez, V. M., Otte, K., Velagapudi, V., Toppari, J., Wilson, D. B. and Heikinheimo, M. (2016) GATA4 Regulates Blood-Testis Barrier Function and Lactate Metabolism in Mouse Sertoli Cells. Endocrinology 157(6): 2416-31. PMID: [26974005]
Schrade, A., Kyronlahti, A., Akinrinade, O., Pihlajoki, M., Fischer, S., Rodriguez, V. M., Otte, K., Velagapudi, V., Toppari, J., Wilson, D. B. and Heikinheimo, M. (2016) GATA4 Regulates Blood-Testis Barrier Function and Lactate Metabolism in Mouse Sertoli Cells. Endocrinology 157(6): 2416-31. PMID: [26974005]
 Schrade, A., Kyronlahti, A., Akinrinade, O., Pihlajoki, M., Fischer, S., Rodriguez, V. M., Otte, K., Velagapudi, V., Toppari, J., Wilson, D. B. and Heikinheimo, M. (2016) GATA4 Regulates Blood-Testis Barrier Function and Lactate Metabolism in Mouse Sertoli Cells. Endocrinology 157(6): 2416-31. PMID: [26974005]
Schrade, A., Kyronlahti, A., Akinrinade, O., Pihlajoki, M., Fischer, S., Rodriguez, V. M., Otte, K., Velagapudi, V., Toppari, J., Wilson, D. B. and Heikinheimo, M. (2016) GATA4 Regulates Blood-Testis Barrier Function and Lactate Metabolism in Mouse Sertoli Cells. Endocrinology 157(6): 2416-31. PMID: [26974005]
 Schrade, A., Kyronlahti, A., Akinrinade, O., Pihlajoki, M., Fischer, S., Rodriguez, V. M., Otte, K., Velagapudi, V., Toppari, J., Wilson, D. B. and Heikinheimo, M. (2016) GATA4 Regulates Blood-Testis Barrier Function and Lactate Metabolism in Mouse Sertoli Cells. Endocrinology 157(6): 2416-31. PMID: [26974005]
Schrade, A., Kyronlahti, A., Akinrinade, O., Pihlajoki, M., Fischer, S., Rodriguez, V. M., Otte, K., Velagapudi, V., Toppari, J., Wilson, D. B. and Heikinheimo, M. (2016) GATA4 Regulates Blood-Testis Barrier Function and Lactate Metabolism in Mouse Sertoli Cells. Endocrinology 157(6): 2416-31. PMID: [26974005]
 Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
 Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
 Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
 Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
 Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
 Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
 Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
 Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
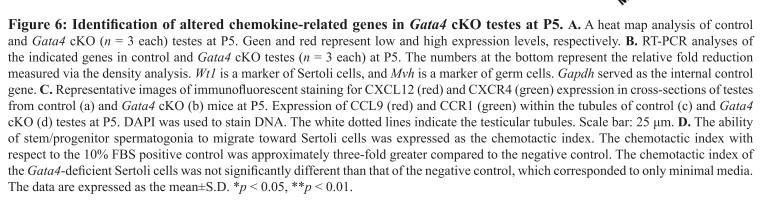 Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
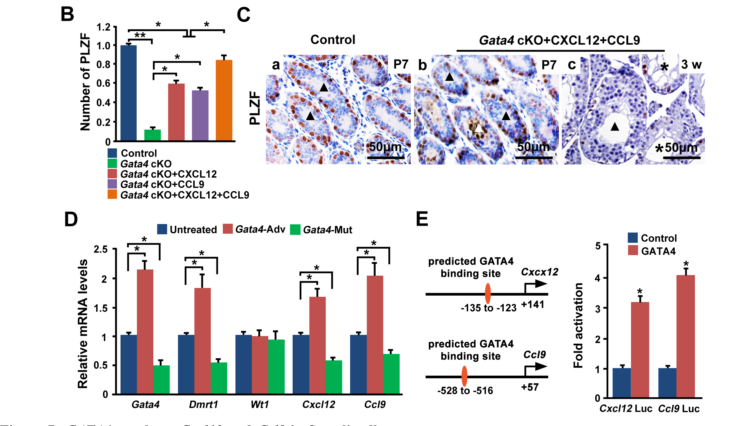 Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
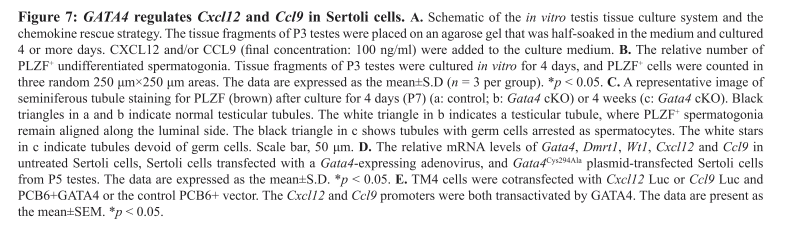 Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]
Chen, S. R., Tang, J. X., Cheng, J. M., Li, J., Jin, C., Li, X. Y., Deng, S. L., Zhang, Y., Wang, X. X. and Liu, Y. X. (2015) Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 6(35): 37012-27. PMID: [26473289]















