General information
| SG000351 | |||||||||||||||||
| Hsp90ab1 | |||||||||||||||||
| 90kDa, AL022974, C81438, Hsp84, Hsp84-1, Hsp90, Hspcb | |||||||||||||||||
| Mus musculus | |||||||||||||||||
| 10090 | |||||||||||||||||
| ENSMUSG00000023944 | |||||||||||||||||
| ENSMUSP00000024739 ENSMUSP00000126239 ENSMUSP00000127338 ENSMUSP00000131601 ENSMUSP00000119678 | |||||||||||||||||
Heat shock protein 90 alpha (cytosolic), class B member 1 [Mus musculus(house mouse)] | |||||||||||||||||
| |||||||||||||||||
Reviewed functional gene |
Functional information
| premeiotic meiotic | ||||||||||||||||
| Spermatocyte Spermatogonia | ||||||||||||||||
|
1. Abstract | ||||||||||||||||
| ||||||||||||||||
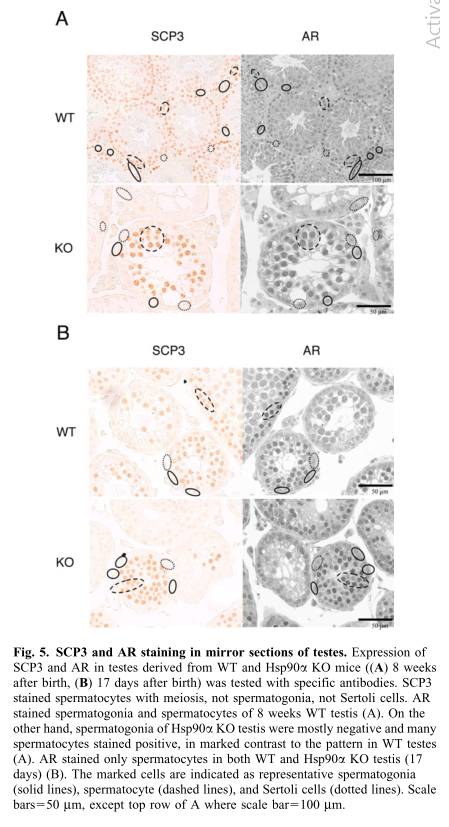
| AR,specifically chaperoned by Hsp90-alpha in spermatogonia, is critical for maintenance of established spermatogenesis and for survival of spermatocytes in adult testis,in addition to setting the first wave of spermatogenesis before puberty | ||||||||||||||||
|
Click the right sign for more information
| ||||||||||||||||
|
ReactomeID: R-MMU-8937144 Aryl hydrocarbon receptor signalling | ||||||||||||||||
| N/A |
Expression and location
| Expressed highest in ovary | |
| Expressed highest in spermatogonium | |
| View detail | |
| View detail | |
| Cytoplasm |
Mutation in human orthology
|
1000G (Phase 3): ESP6500 (SI-V2): ExAC (r0.3.1): dbSNP(Build 147): |
|||||||
|
Chinese health control (254): European health control (283): Chinese patients (168): |
|||||||
|
Other information
|
Click the right sign for more information
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| P11499 |