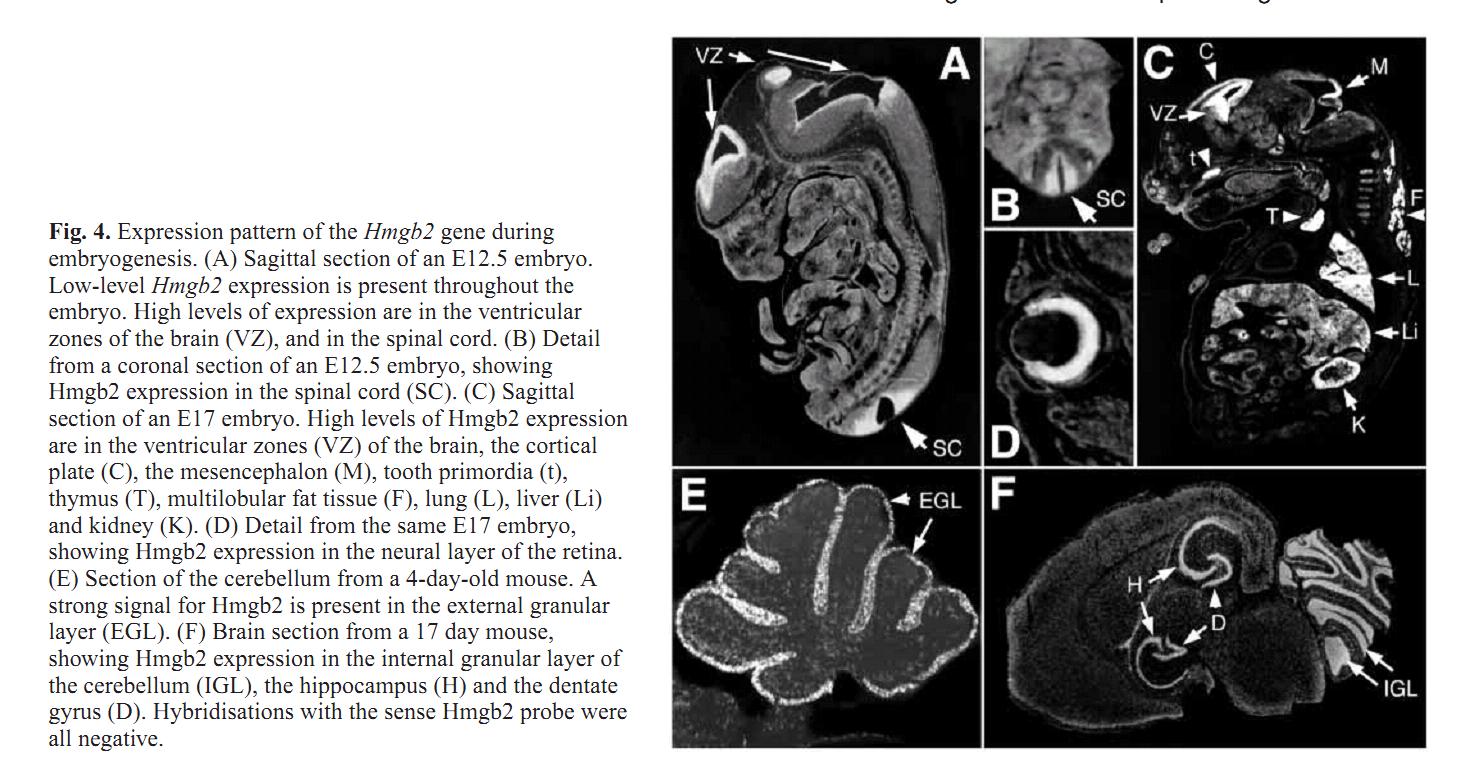
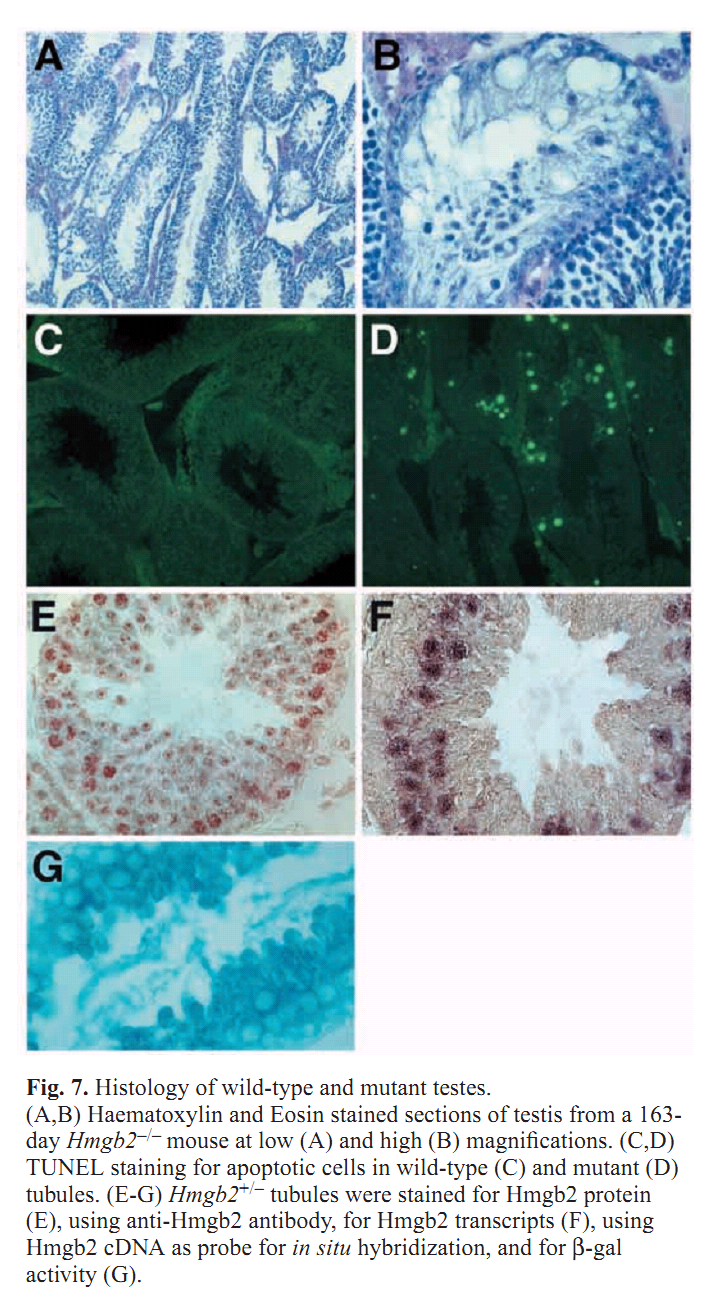
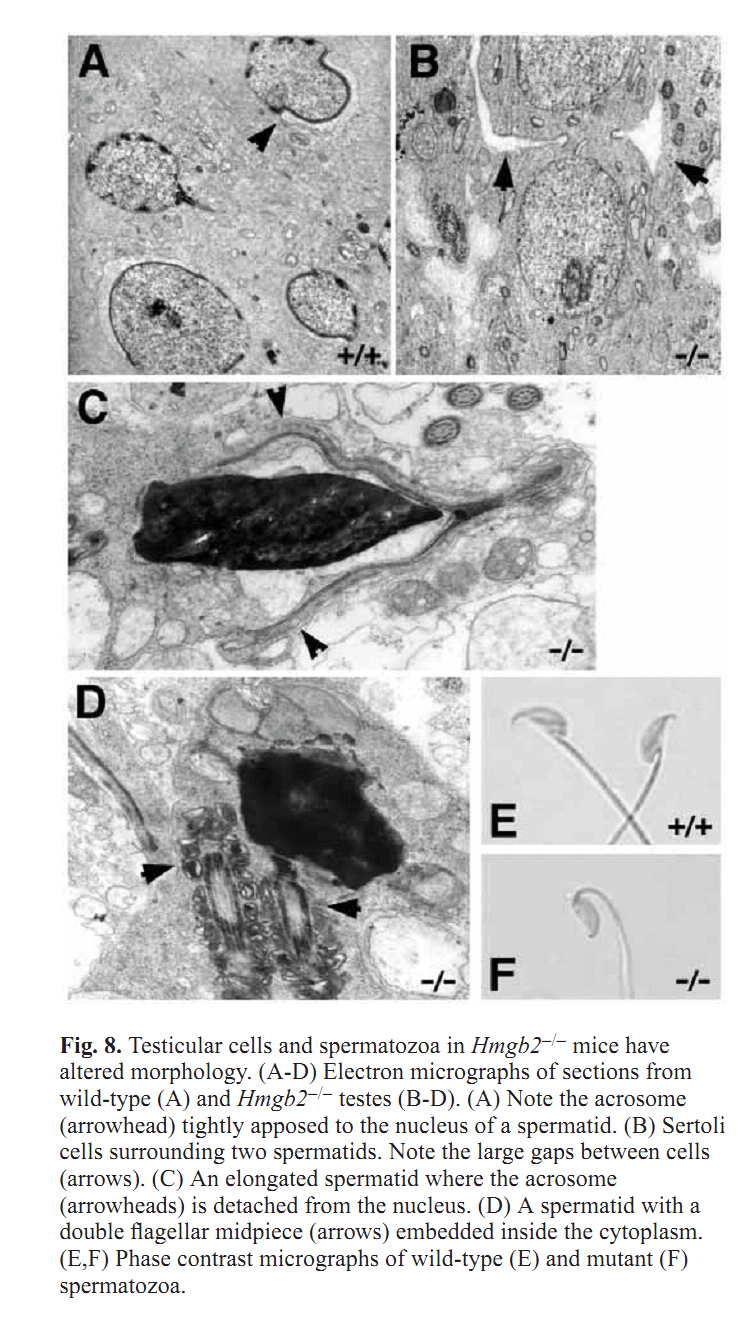
| Sub-Ontologies | Accession | Term | Evidence |
| Biological Process | GO:0001938 | positive regulation of endothelial cell proliferation | IEA |
| Biological Process | GO:0002376 | immune system process | IEA |
| Biological Process | GO:0002437 | inflammatory response to antigenic stimulus | IMP |
| Biological Process | GO:0006265 | DNA topological change | ISS |
| Biological Process | GO:0006310 | DNA recombination | IEA |
| Biological Process | GO:0006351 | transcription, DNA-templated | IEA |
| Biological Process | GO:0006355 | regulation of transcription, DNA-templated | IEA |
| Biological Process | GO:0006357 | regulation of transcription from RNA polymerase II promoter | IEA |
| Biological Process | GO:0006935 | chemotaxis | IEA |
| Biological Process | GO:0006954 | inflammatory response | IEA |
| Biological Process | GO:0007283 | spermatogenesis | IMP |
| Biological Process | GO:0007289 | spermatid nucleus differentiation | IMP |
| Biological Process | GO:0008584 | male gonad development | IMP |
| Biological Process | GO:0010628 | positive regulation of gene expression | IDA |
| Biological Process | GO:0010629 | negative regulation of gene expression | IDA |
| Biological Process | GO:0032075 | positive regulation of nuclease activity | IEA |
| Biological Process | GO:0032496 | response to lipopolysaccharide | IMP |
| Biological Process | GO:0032728 | positive regulation of interferon-beta production | IMP |
| Biological Process | GO:0043388 | positive regulation of DNA binding | IEA |
| Biological Process | GO:0045087 | innate immune response | IEA |
| Biological Process | GO:0045089 | positive regulation of innate immune response | IMP |
| Biological Process | GO:0045648 | positive regulation of erythrocyte differentiation | IEA |
| Biological Process | GO:0045654 | positive regulation of megakaryocyte differentiation | IEA |
| Biological Process | GO:0045892 | negative regulation of transcription, DNA-templated | IEA |
| Biological Process | GO:0045893 | positive regulation of transcription, DNA-templated | IEA |
| Biological Process | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA |
| Biological Process | GO:0048545 | response to steroid hormone | IMP |
| Biological Process | GO:0050767 | regulation of neurogenesis | IMP |
| Biological Process | GO:0050829 | defense response to Gram-negative bacterium | IEA |
| Biological Process | GO:0050830 | defense response to Gram-positive bacterium | IEA |
| Biological Process | GO:0050918 | positive chemotaxis | IEA |
| Biological Process | GO:0060326 | cell chemotaxis | IEA |
| Biological Process | GO:0071222 | cellular response to lipopolysaccharide | IEA |
| Biological Process | GO:0072091 | regulation of stem cell proliferation | IMP |
| Biological Process | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | IMP |
| Cellular Component | GO:0000790 | nuclear chromatin | IDA |
| Cellular Component | GO:0000793 | condensed chromosome | IEA |
| Cellular Component | GO:0005576 | extracellular region | IEA |
| Cellular Component | GO:0005615 | extracellular space | IDA |
| Cellular Component | GO:0005623 | cell | IEA |
| Cellular Component | GO:0005634 | nucleus | IDA |
| Cellular Component | GO:0005694 | chromosome | IEA |
| Cellular Component | GO:0005730 | nucleolus | IEA |
| Cellular Component | GO:0005737 | cytoplasm | IDA |
| Cellular Component | GO:0043234 | protein complex | IEA |
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | IEA |
| Molecular Function | GO:0001158 | enhancer sequence-specific DNA binding | IDA |
| Molecular Function | GO:0003677 | DNA binding | IEA |
| Molecular Function | GO:0003684 | damaged DNA binding | IEA |
| Molecular Function | GO:0003690 | double-stranded DNA binding | IDA |
| Molecular Function | GO:0003700 | transcription factor activity, sequence-specific DNA binding | IEA |
| Molecular Function | GO:0003723 | RNA binding | IEA |
| Molecular Function | GO:0008134 | transcription factor binding | IDA |
| Molecular Function | GO:0008301 | DNA binding, bending | IEA |
| Molecular Function | GO:0019904 | protein domain specific binding | IPI |
| Molecular Function | GO:0042056 | chemoattractant activity | IEA |
| Molecular Function | GO:0044212 | transcription regulatory region DNA binding | IEA |
| Molecular Function | GO:0044378 | non-sequence-specific DNA binding, bending | ISS |
| Molecular Function | GO:0050786 | RAGE receptor binding | IEA |
| Molecular Function | GO:0097100 | supercoiled DNA binding | ISS |