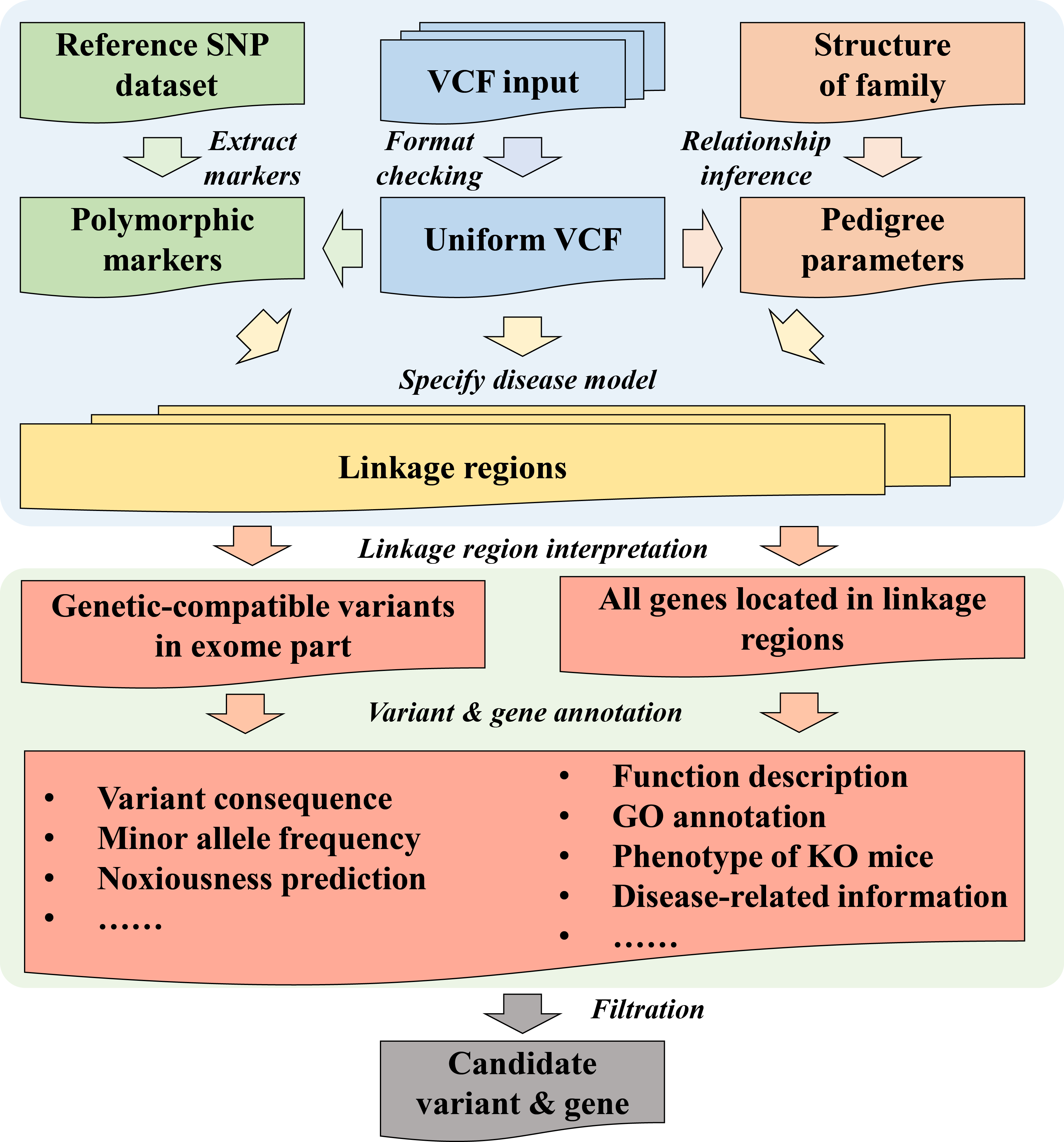
The advance in Next Generation Sequencing technology has revolutionized the field of disease research. However, pinpointing the disease-causing variants from millions of variants is still a tough job. Here, based on a review of traditional linkage analysis tools, we present PedMiner, a web-based application designed to perform linkage analysis directly using family-based whole-exome sequencing data to narrow down the candidate variants. PedMiner uses standard Variant Call Format files (VCF) as input, together with its graphical interface design, making it an easy-to-use application. PedMiner integrates linkage analysis, variant annotation, and variant prioritization to a one-step automated pipeline. It provides graphical visualization of the linkage region and comprehensive annotation of variants/genes within the linkage region. This efficient and comprehensive application will make ease for the scientific community working on Mendelian diseases based on family-based whole-exome sequencing data.
The server is implemented in PHP + python + R script and is online available to all users for free at http://mcg.ustc.edu.cn/bsc/pedminer/. If you have any problem, please feel free to contact us.